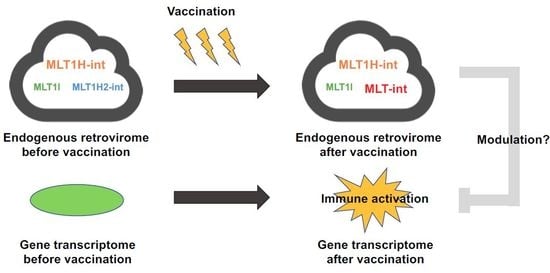
Identification of a Retroelement-Containing Human Transcript Induced in the Nucleus by Vaccination
Abstract
:1. Introduction
2. Results
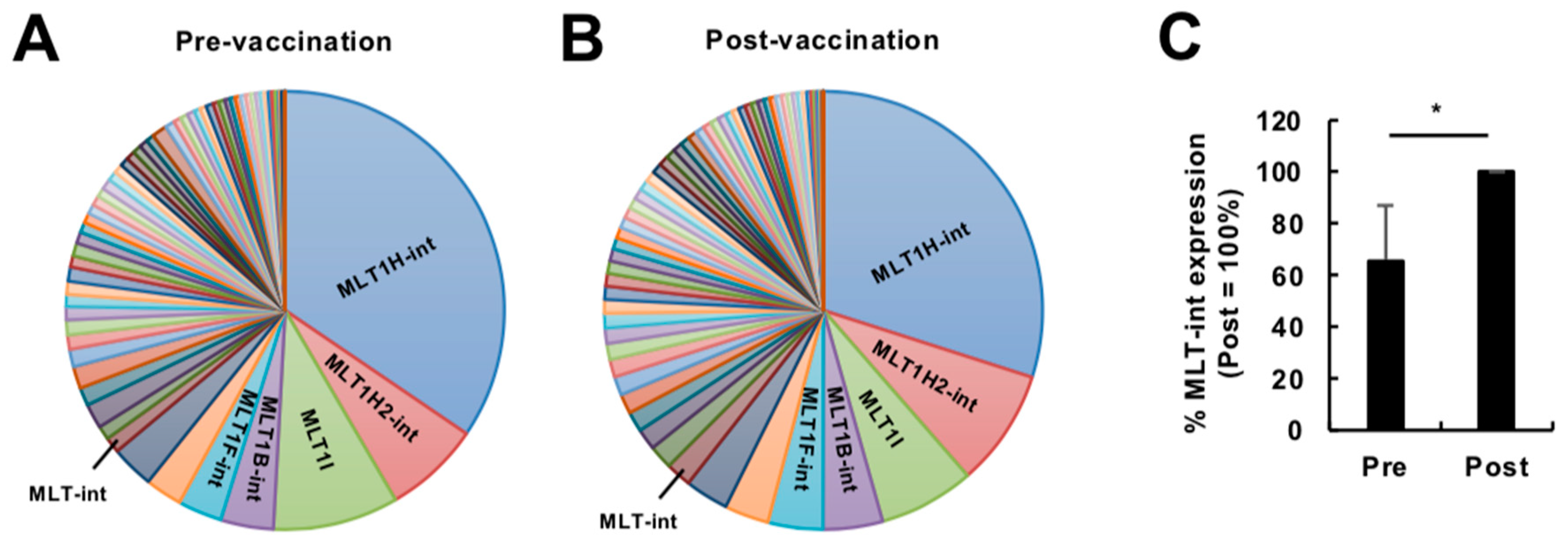
2.1. Expression Profiling of Retroelement-Containing Transcripts in the Human Blood during Vaccination
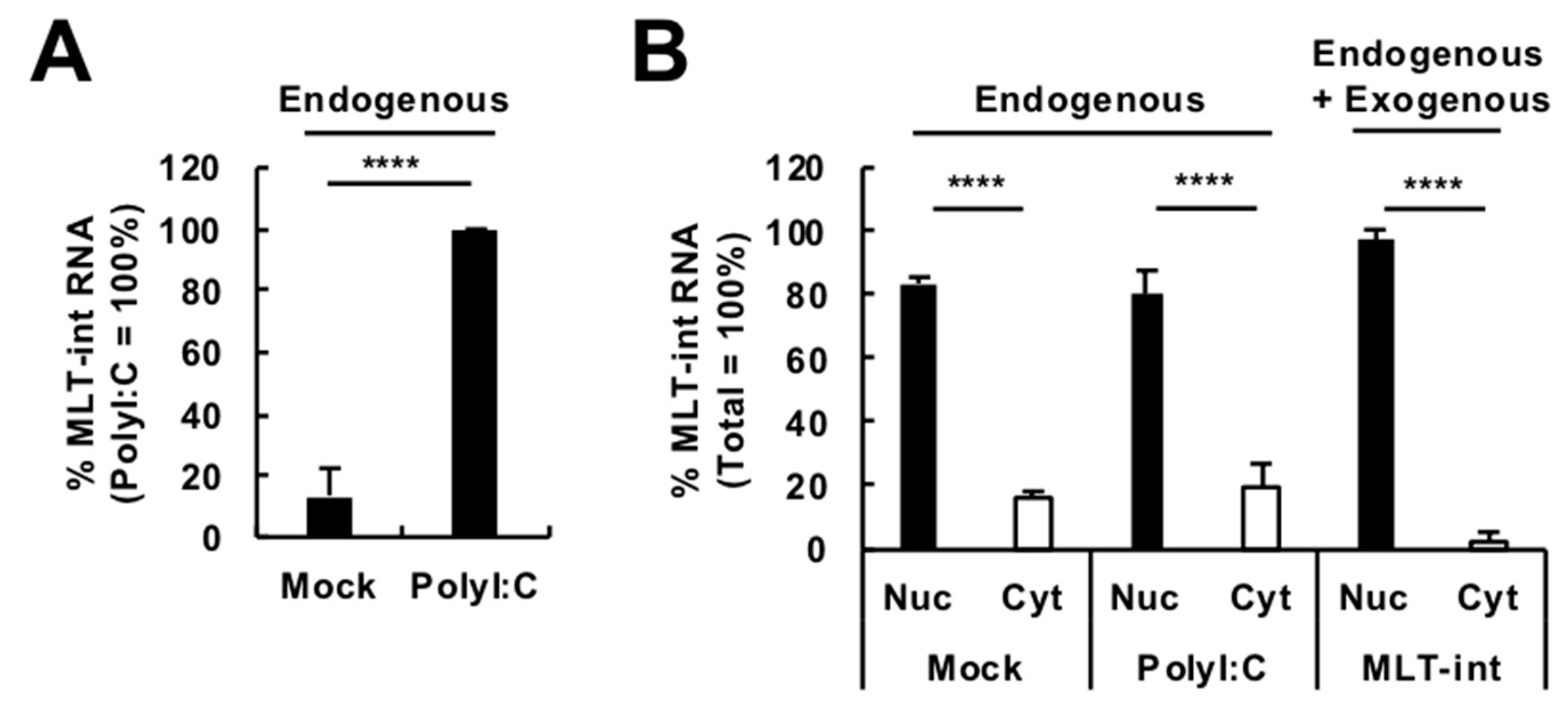
2.2. Localization of MLT-int-Containing Transcripts in the Nucleus
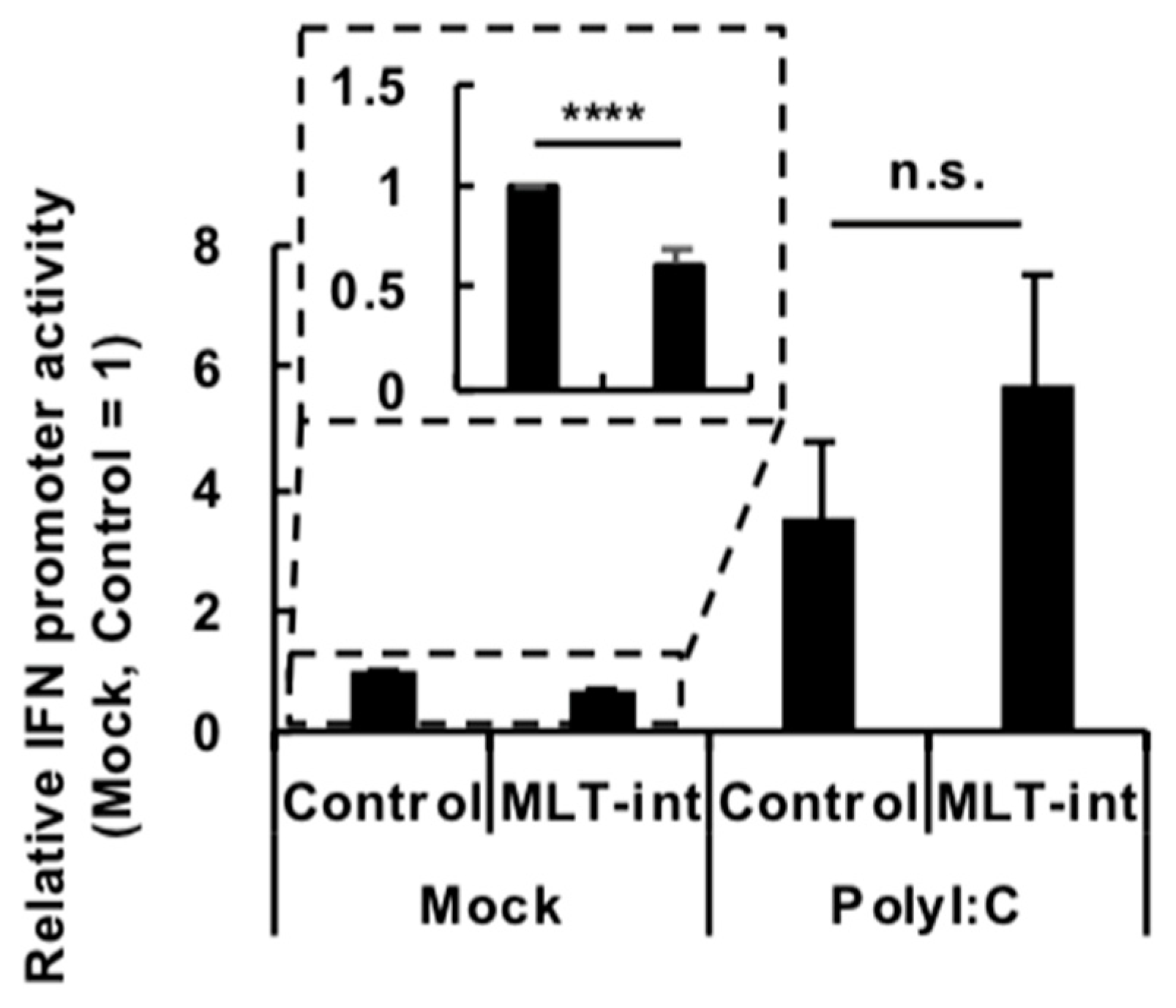
2.3. Suppression of the IFN Promoter Activity by MLT-int in the Absence of Immune Stimuli
3. Discussion
4. Materials and Methods
4.1. RNA-seq Analysis
4.2. Plasmids
4.3. Primers
4.4. Cells
4.5. Subcellular Fractionation
4.6. Luciferase Reporter Assay
4.7. Real-Time PCR
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ERV | endogenous retrovirus |
| IFN | interferon |
| ISG | IFN-stimulated gene |
| ISRE | IFN-stimulated response element |
| LINE | long interspersed element |
| MaLR | mammalian apparent LTR-retrotransposon |
| ncRNA | non-coding RNA |
| RIG-I | retinoic acid-inducible gene I |
| SINE | short interspersed element |
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mager, D.L.; Stoye, J.P. Mammalian endogenous retroviruses. Microbiol. Spectr. 2014, 3, 1–20. [Google Scholar] [CrossRef]
- Kassiotis, G.; Stoye, J.P. Making a virtue of necessity: The pleiotropic role of human endogenous retroviruses in cancer. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160277. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Attig, J.; Young, G.R.; Stoye, J.P.; Kassiotis, G. Physiological and pathological transcriptional activation of endogenous retroelements assessed by RNA-sequencing of B lymphocytes. Front. Microbiol. 2017, 8, 2489. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.; Mistry, A.; Keding, A.; Gabe, R.; Cook, E.; Forrester, S.; Wiggins, R.; Di Marco, S.; Colloca, S.; Siani, L.; et al. A third generation vaccine for human visceral leishmaniasis and post kala azar dermal leishmaniasis: First-in-human trial of ChAd63-KH. PLoS Negl. Trop. Dis. 2017, 11, e0005527. [Google Scholar] [CrossRef]
- Jurka, J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000, 16, 418–420. [Google Scholar] [CrossRef]
- Smit, A.F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993, 21, 1863–1872. [Google Scholar] [CrossRef] [Green Version]
- Kojima, K.K. Human transposable elements in Repbase: Genomic footprints from fish to humans. Mob. DNA 2018, 9, 2. [Google Scholar] [CrossRef]
- Wong, J.J.Y.; Pung, Y.F.; Sze, N.S.-K.; Chin, K.-C. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. USA 2006, 103, 10735–10740. [Google Scholar] [CrossRef] [Green Version]
- Kassiotis, G.; Stoye, J.P. Immune responses to endogenous retroelements: Taking the bad with the good. Nat. Rev. Immunol. 2016, 16, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Sachs, F.; Ramsay, L.; Jacques, P.-É.; Göke, J.; Bourque, G.; Ng, H.-H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 2014, 21, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebert, P.J.R.; Jiang, S.; Xie, J.; Li, Q.-J.; Davis, M.M. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat. Immunol. 2009, 10, 1162–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Q.; Carbone, C.J.; Katlinskaya, Y.V.; Zheng, H.; Zheng, K.; Luo, M.; Wang, P.J.; Greenberg, R.A.; Fuchs, S.Y. Type I interferon controls propagation of long interspersed element-1. J. Biol. Chem. 2015, 290, 10191–10199. [Google Scholar] [CrossRef] [PubMed]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Honda, T.; Tomonaga, K. Endogenous non-retroviral RNA virus elements evidence a novel type of antiviral immunity. Mob. Genet. Elem. 2016, 6, e1165785. [Google Scholar] [CrossRef] [Green Version]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef] [Green Version]
- Best, S.; Le Tissier, P.; Towers, G.; Stoye, J.P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 1996, 382, 826–829. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Takemoto, K.; Ueda, K.; Dividsion of Virology, Department of Microbiology and Immunology, Osaka University Graduate School of Medicine, Suita, Osaka, Japan. MLT-int RNA was not detected in the RIG-I immunoprecipitates. 2019. [Google Scholar]
- Shaw, A.E.; Hughes, J.; Gu, Q.; Behdenna, A.; Singer, J.B.; Dennis, T.; Orton, R.J.; Varela, M.; Gifford, R.J.; Wilson, S.J.; et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017, 15, e2004086. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneyama, M.; Suhara, W.; Fukuhara, Y.; Sato, M.; Ozato, K.; Fujita, T. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3). J. Biochem. 1996, 120, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, Y.; Nakayama, R.; Obika, S.; Ohsaki, E.; Ueda, K.; Honda, T. Inhibition of LINE-1 Retrotransposition by Capsaicin. Int. J. Mol. Sci. 2018, 19, 3243. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, R.; Ueno, Y.; Ueda, K.; Honda, T. Latent infection with Kaposi’s sarcoma-associated herpesvirus enhances retrotransposition of long interspersed element-1. Oncogene 2019, 38, 4340–4351. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Honda, T.; Ueda, K. vFLIP upregulates IKKε, leading to spindle morphology formation through RelA activation. Virology 2018, 522, 106–121. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Honda, T.; Takemoto, K.; Ueda, K. Identification of a Retroelement-Containing Human Transcript Induced in the Nucleus by Vaccination. Int. J. Mol. Sci. 2019, 20, 2875. https://doi.org/10.3390/ijms20122875
Honda T, Takemoto K, Ueda K. Identification of a Retroelement-Containing Human Transcript Induced in the Nucleus by Vaccination. International Journal of Molecular Sciences. 2019; 20(12):2875. https://doi.org/10.3390/ijms20122875
Chicago/Turabian StyleHonda, Tomoyuki, Keiko Takemoto, and Keiji Ueda. 2019. "Identification of a Retroelement-Containing Human Transcript Induced in the Nucleus by Vaccination" International Journal of Molecular Sciences 20, no. 12: 2875. https://doi.org/10.3390/ijms20122875