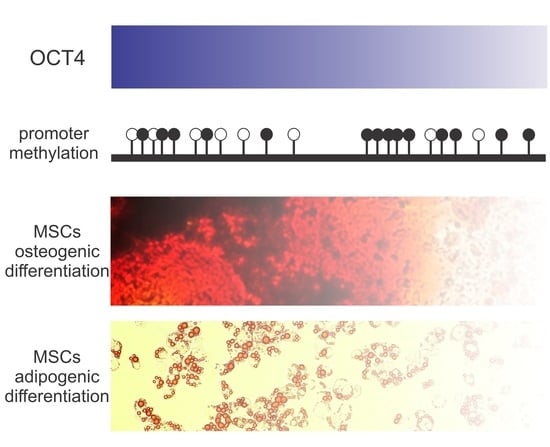
OCT4 Silencing Triggers Its Epigenetic Repression and Impairs the Osteogenic and Adipogenic Differentiation of Mesenchymal Stromal Cells
Abstract
:1. Introduction
2. Results
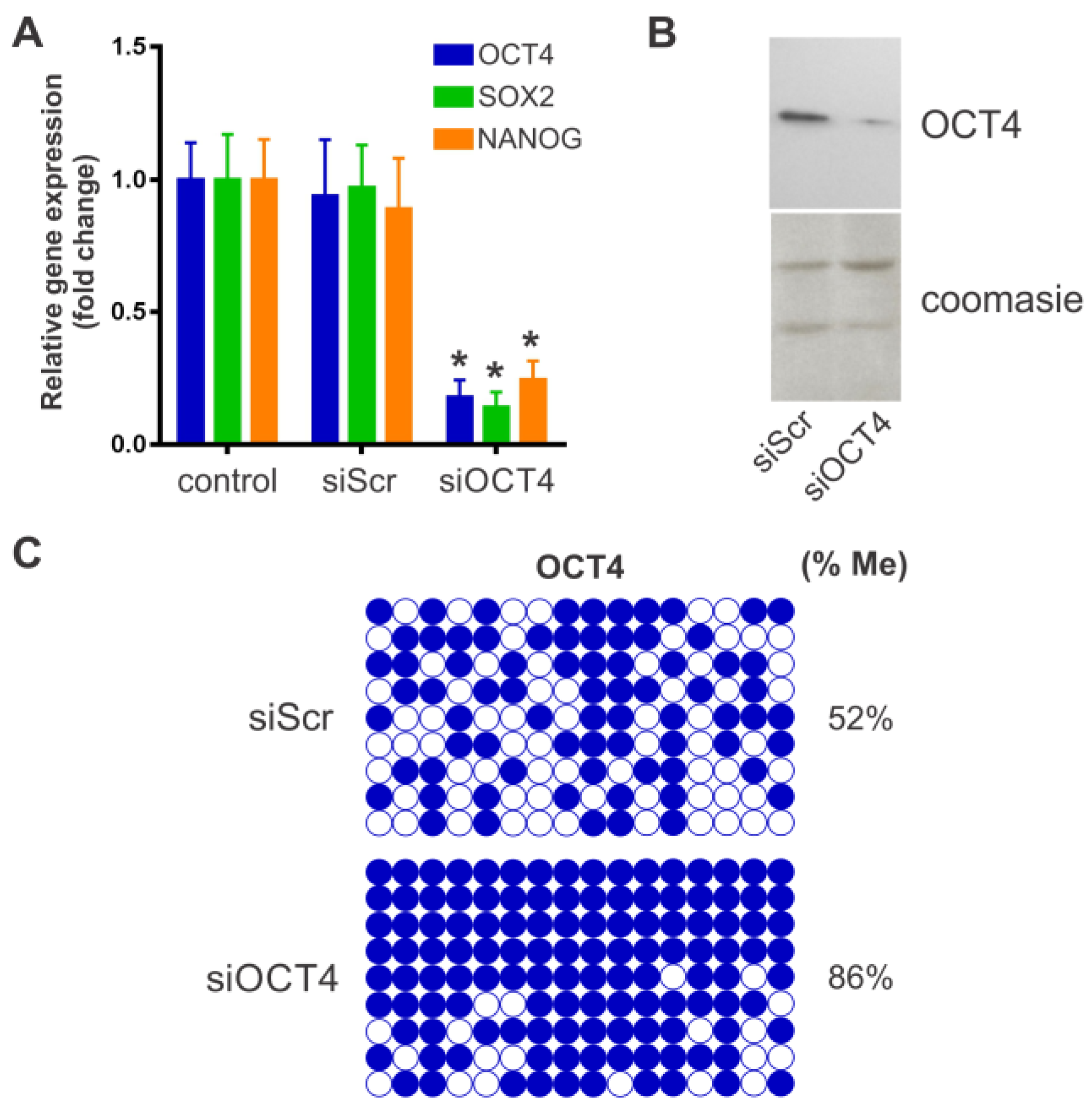
2.1. BM-MSC Characterization and OCT4 Silencing
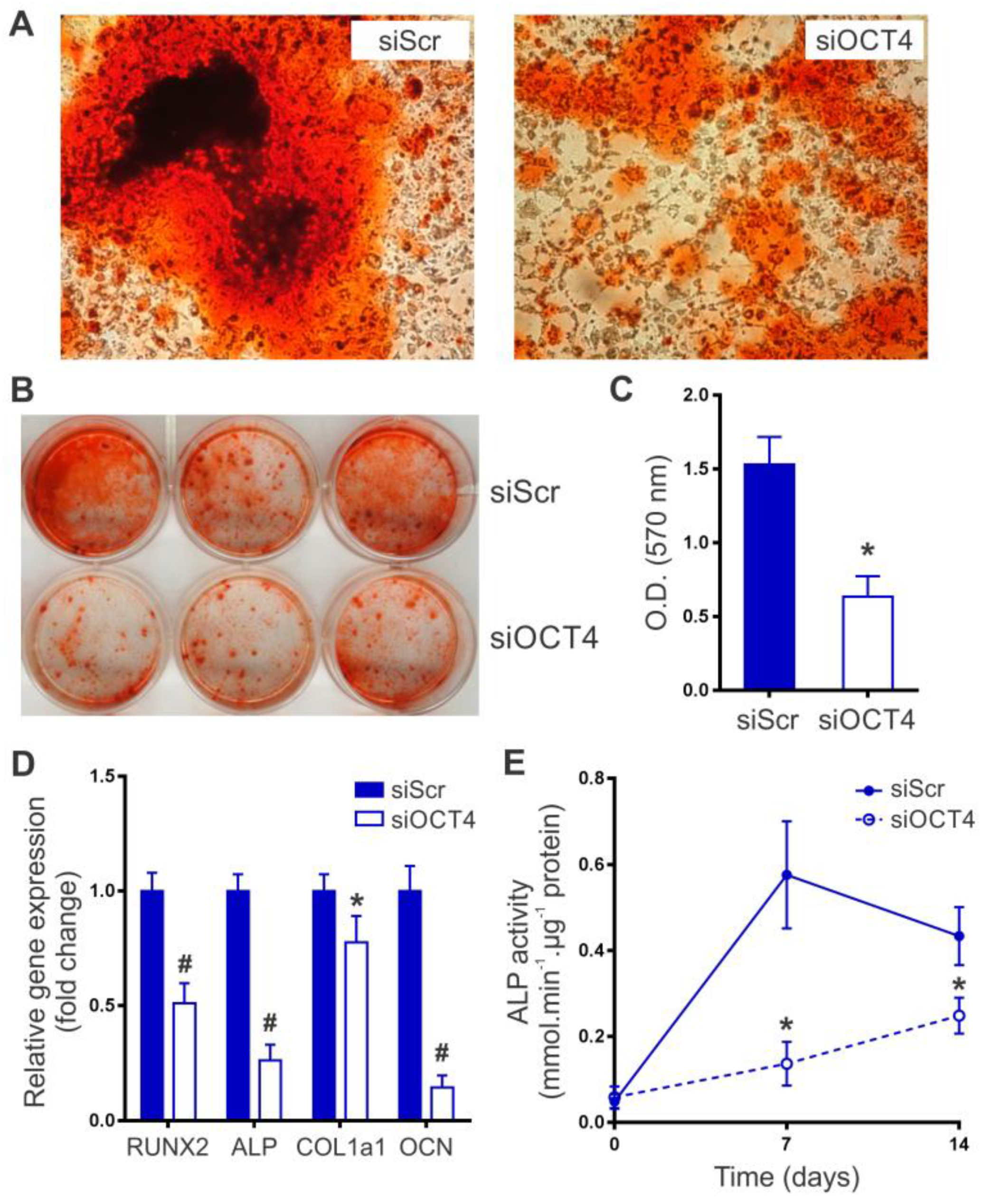
2.2. OCT4 Silencing Impairs MSC Osteogenic Differentiation
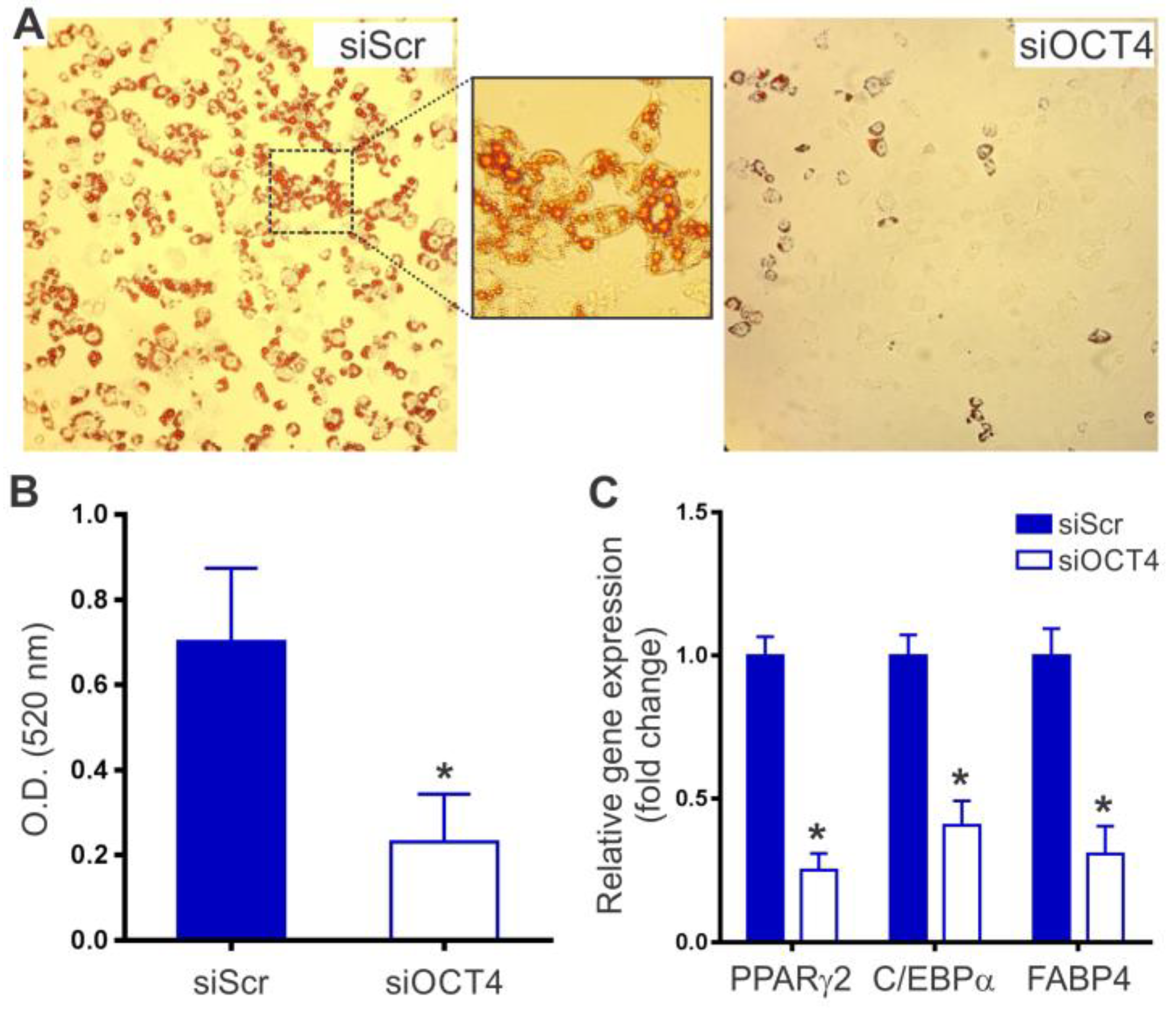
2.3. OCT4 Silencing Prevents MSC Adipocyte Differentiation
3. Discussion
4. Materials and Methods
4.1. Bone Marrow-Derived Mesenchymal Stromal Cells
4.2. OCT4 Silencing
4.3. Quantitative RT-PCR
4.4. Bisulfite Genomic Sequencing
4.5. Western Blotting
4.6. Osteogenic Differentiation
4.7. Adipogenic Differentiation
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- Silva, J.; Smith, A. Capturing pluripotency. Cell 2008, 132, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Tantin, D. Oct transcription factors in development and stem cells: Insights and mechanisms. Development 2013, 140, 2857–2866. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Sterneckert, J.; Höing, S.; Schöler, H.R. Concise review: Oct4 and more: The reprogramming expressway. Stem Cells 2012, 30, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Miyazaki, J.; Smith, A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000, 24, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Simandi, Z.; Horvath, A.; Wright, L.C.; Cuaranta-Monroy, I.; De Luca, I.; Karolyi, K.; Sauer, S.; Deleuze, J.F.; Gudas, L.J.; Cowley, S.M.; et al. OCT4 acts as an integrator of pluripotency and signal-induced differentiation. Mol. Cell 2016, 63, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Colter, D.C.; Class, R.; DiGirolamo, C.M.; Prockop, D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc. Natl. Acad. Sci. USA 2000, 97, 3213–3218. [Google Scholar] [CrossRef] [PubMed]
- Pochampally, R.R.; Smith, J.R.; Ylostalo, J.; Prockop, D.J. Serum deprivation of human marrow stromal cells (hMSCs) selects for a subpopulation of early progenitor cells with enhanced expression of OCT-4 and other embryonic genes. Blood 2004, 103, 1647–1652. [Google Scholar] [CrossRef] [Green Version]
- Yannarelli, G.; Pacienza, N.; Cuniberti, L.; Medin, J.; Davies, J.; Keating, A. Brief report: The potential role of epigenetics on multipotent cell differentiation capacity of mesenchymal stromal cells. Stem Cells 2013, 31, 215–220. [Google Scholar] [CrossRef]
- Yannarelli, G.; Dayan, V.; Pacienza, N.; Lee, C.J.; Medin, J.; Keating, A. Human umbilical cord perivascular cells exhibit enhanced cardiomyocyte reprogramming and cardiac function after experimental acute myocardial infarction. Cell Transplant. 2013, 22, 1651–1666. [Google Scholar] [CrossRef]
- Hao, Q.; An, J.Q.; Hao, F.; Yang, C.; Lu, T.; Qu, T.Y.; Zhao, L.R.; Duan, W.M. Inducible Lentivirus-Mediated Expression of the Oct4 Gene Affects Multilineage Differentiation of Adult Human Bone Marrow-Derived Mesenchymal Stem Cells. Cell. Reprogram. 2015, 17, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.M.; Wu, Y.N.; Guo, X.M.; Hui, J.H.; Lee, E.H.; Lim, B. Effects of ectopic Nanog and Oct4 overexpression on mesenchymal stem cells. Stem Cells Dev. 2009, 18, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; Su, P.F.; Huang, Y.F.; Yew, T.L.; Hung, S.C. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol. Cell 2012, 47, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Yannarelli, G.; Pacienza, N.; Montanari, S.; Santa-Cruz, D.; Viswanathan, S.; Keating, A. OCT4 expression mediates partial cardiomyocyte reprogramming of mesenchymal stromal cells. PLoS ONE 2017, 12, e0189131. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Xu, L.; Chen, R.; Zhang, T.; Lin, S.; Hou, Y.; Liu, Y.; Meng, F.; Liu, Z.; Ni, M.; et al. Epigenetic memory gained by priming with osteogenic induction medium improves osteogenesis and other properties of mesenchymal stem cells. Sci. Rep. 2015, 5, 11056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef]
- Orimo, H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J. Nippon Med. Sch. 2010, 77, 4–12. [Google Scholar] [CrossRef]
- Ramírez-Zacarías, J.L.; Castro-Muñozledo, F.; Kuri-Harcuch, W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 1992, 97, 493–497. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef]
- Palma, C.S.; Tannous, M.A.; Malta, T.M.; Russo, E.M.; Covas, D.T.; Picanço-Castro, V. Forced expression of OCT4 influences the expression of pluripotent genes in human mesenchymal stem cells and fibroblasts. Genet. Mol. Res. 2013, 12, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Rompolas, P.; Mesa, K.R.; Greco, V. Spatial organization within a niche as a determinant of stem-cell fate. Nature 2013, 502, 513–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altun, G.; Loring, J.F.; Laurent, L.C. DNA methylation in embryonic stem cells. J. Cell. Biochem. 2010, 109, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chew, J.L.; Loh, Y.H.; Zhang, W.; Chen, X.; Tam, W.L.; Yeap, L.S.; Li, P.; Ang, Y.S.; Lim, B.; Robson, P.; et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005, 25, 6031–6046. [Google Scholar] [CrossRef] [PubMed]
- Rodda, D.J.; Chew, J.L.; Lim, L.H.; Loh, Y.H.; Wang, B.; Ng, H.H.; Robson, P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005, 280, 24731–24737. [Google Scholar] [CrossRef]
- Go, M.J.; Takenaka, C.; Ohgushi, H. Forced expression of Sox2 or Nanog in human bone marrow derived mesenchymal stem cells maintains their expansion and differentiation capabilities. Exp. Cell Res. 2008, 314, 1147–1154. [Google Scholar] [CrossRef]
- Marthaler, A.G.; Adachi, K.; Tiemann, U.; Wu, G.; Sabour, D.; Velychko, S.; Kleiter, I.; Schöler, H.R.; Tapia, N. Enhanced OCT4 transcriptional activity substitutes for exogenous SOX2 in cellular reprogramming. Sci. Rep. 2016, 6, 19415. [Google Scholar] [CrossRef] [Green Version]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef]
- Moroni, L.; Fornasari, P.M. Human mesenchymal stem cells: A bank perspective on the isolation, characterization and potential of alternative sources for the regeneration of musculoskeletal tissues. J. Cell. Physiol. 2013, 228, 680–687. [Google Scholar] [CrossRef]
- Kwon, A.; Kim, Y.; Kim, M.; Kim, J.; Choi, H.; Jekarl, D.W.; Lee, S.; Kim, J.M.; Shin, J.C.; Park, I.Y. Tissue-specific Differentiation Potency of Mesenchymal Stromal Cells from Perinatal Tissues. Sci. Rep. 2016, 6, 23544. [Google Scholar] [CrossRef]
- Ramkisoensing, A.A.; Pijnappels, D.A.; Askar, S.F.; Passier, R.; Swildens, J.; Goumans, M.J.; Schutte, C.I.; de Vries, A.A.; Scherjon, S.; Mummery, C.L.; et al. Human embryonic and fetal mesenchymal stem cells differentiate toward three different cardiac lineages in contrast to their adult counterparts. PLoS ONE 2011, 6, e24164. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, E.; Koh, S.E.; Maeng, S.; Lee, W.D.; Lim, J.; Shim, I.; Lee, Y.J. Dopaminergic differentiation of neural progenitors derived from placental mesenchymal stem cells in the brains of Parkinson’s disease model rats and alleviation of asymmetric rotational behavior. Brain Res. 2012, 1466, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Boroujeni, M.E.; Gardaneh, M. Umbilical cord: An unlimited source of cells differentiable towards dopaminergic neurons. Neural Regen. Res. 2017, 12, 1186–1192. [Google Scholar] [PubMed]
- Jiang, S.; Zhang, S. Differentiation of cardiomyocytes from amniotic fluid‑derived mesenchymal stem cells by combined induction with transforming growth factor β1 and 5‑azacytidine. Mol. Med. Rep. 2017, 16, 5887–5893. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef]
- Saxe, J.P.; Tomilin, A.; Schöler, H.R.; Plath, K.; Huang, J. Post-translational regulation of Oct4 transcriptional activity. PLoS ONE 2009, 4, e4467. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, J.; Li, K.; Wu, T.; Huang, B.; Liu, W.; Kou, X.; Zhang, Y.; Huang, H.; Jiang, Y.; et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 2013, 12, 453–469. [Google Scholar] [CrossRef]
- Rohde, C.; Zhang, Y.; Reinhardt, R.; Jeltsch, A. BISMA—Fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinform. 2010, 11, 230. [Google Scholar] [CrossRef]
- Stanford, C.M.; Jacobson, P.A.; Eanes, E.D.; Lembke, L.A.; Midura, R.J. Rapidly forming apatitic mineral in an osteoblastic cell line (UMR 106-01 BSP). J. Biol. Chem. 1995, 270, 9420–9428. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malvicini, R.; Santa-Cruz, D.; Pacienza, N.; Yannarelli, G. OCT4 Silencing Triggers Its Epigenetic Repression and Impairs the Osteogenic and Adipogenic Differentiation of Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2019, 20, 3268. https://doi.org/10.3390/ijms20133268
Malvicini R, Santa-Cruz D, Pacienza N, Yannarelli G. OCT4 Silencing Triggers Its Epigenetic Repression and Impairs the Osteogenic and Adipogenic Differentiation of Mesenchymal Stromal Cells. International Journal of Molecular Sciences. 2019; 20(13):3268. https://doi.org/10.3390/ijms20133268
Chicago/Turabian StyleMalvicini, Ricardo, Diego Santa-Cruz, Natalia Pacienza, and Gustavo Yannarelli. 2019. "OCT4 Silencing Triggers Its Epigenetic Repression and Impairs the Osteogenic and Adipogenic Differentiation of Mesenchymal Stromal Cells" International Journal of Molecular Sciences 20, no. 13: 3268. https://doi.org/10.3390/ijms20133268