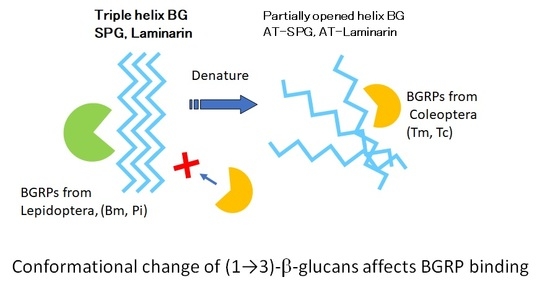
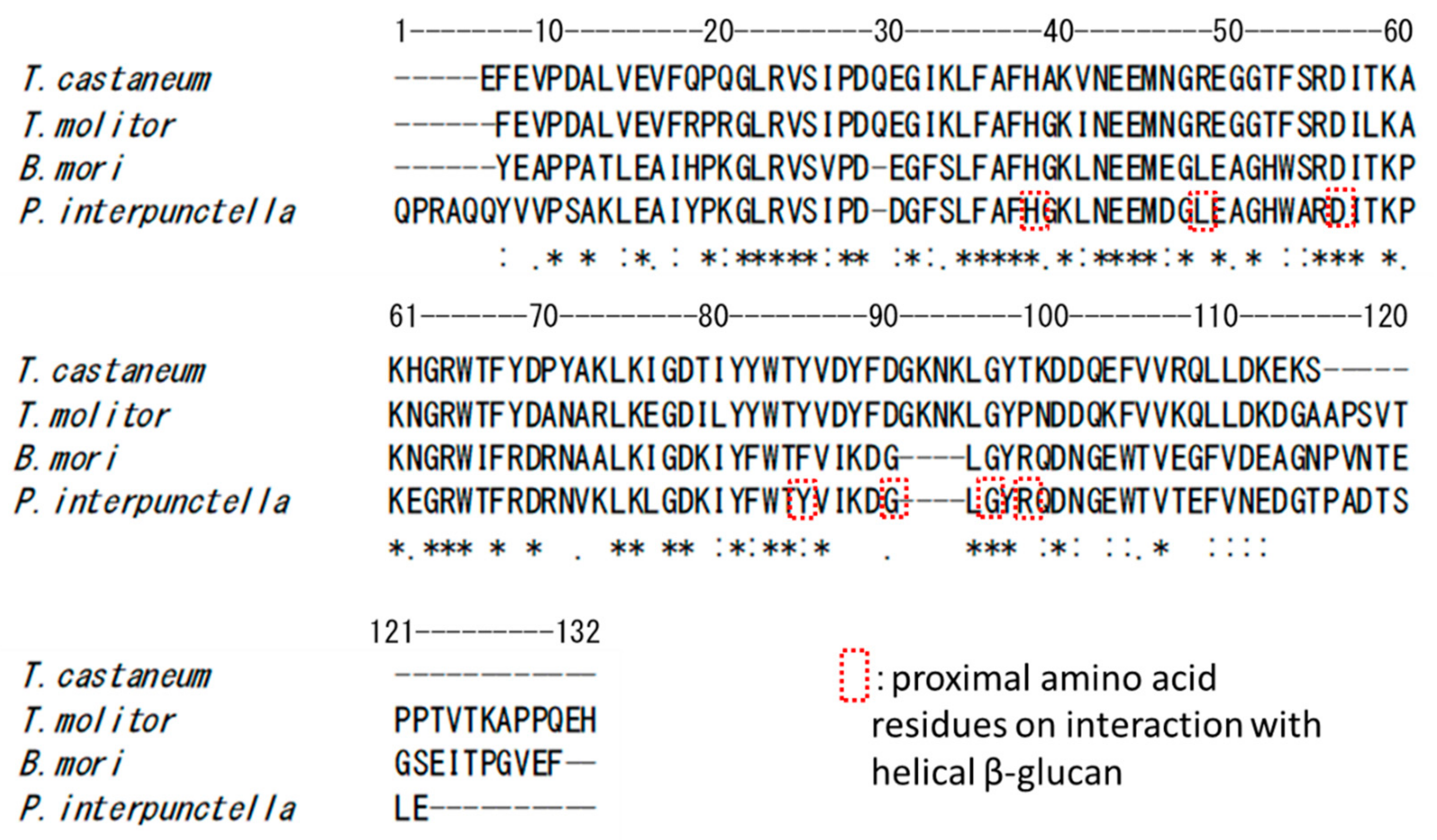
N-Terminal (1→3)-β-d-Glucan Recognition Proteins from Insects Recognize the Difference in Ultra-Structures of (1→3)-β-d-Glucan
Abstract
:1. Introduction
2. Results
2.1. Evaluation of Direct Binding Activity of BGRP-Fc Proteins to Solid Phase BG by ELISA
2.1.1. Binding of BGRPs to Solid Phase of SPG
2.1.2. Binding of BGRPs to Solid Phase of Laminarin
2.2. Competitive Effect of Liquid Phase BGs in ELISA
2.2.1. Binding Difference of BGRPs to the Different Conformation of SPG in Liquid Phase
2.2.2. Binding Difference of BGRPs to the Different Conformation of Laminarin in Liquid Phase
2.3. Binding Kinetics of BGRPs to SPG and AT-SPG
3. Discussion
4. Materials and Methods
4.1. Insect larvae
4.2. β-Glucans
4.3. Preparation of BGRP-Fc Molecules
4.4. Preparation of BGRP-CRD
4.5. Binding Assay of BGRP-Fc to β-Glucans by ELISA
4.6. Competitive ELISA Using Liquid Phase of β-Glucans
4.7. Binding Affinity Studies
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BG | (1→3)-β-d-glucan |
| BGRP | (1→3)-β-d-glucan recognition protein |
| PG | peptidoglycan |
| PGRP | peptidoglycan recognition protein |
| SPG | schizophylan, sonfilan |
| AT | Alkaline-treated |
| Bm | Bombyx mori |
| Pi | Plodia interpunctera |
| Tc | Tribolium castaneum |
| Tm | Tenebrio molita |
| Blitz | biolayer interferometry |
| LAL | Limulus amebocyte lysate |
| SLP | silkworm larvae plasma |
| HRP | horse radish peroxidase |
| TMB | 3,3′,5,5′-tetramethyl-benzidene |
| PBS | phosphate-buffered saline |
| BPBS | PBS containing 0.5% bovine serum albumin |
| PBST | PBS containing 0.05% Tween 20 |
References
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Vetvicka, V.; Sima, P. Various roles of β-glucan in invertebrates. Invertebrate Surviv. J. 2017, 14, 488–493. [Google Scholar]
- Lee, M.H.; Osaki, T.; Lee, J.Y.; Baek, M.J.; Zhang, R.; Park, J.W.; Kawabata, S.; Soderhall, K.; Lee, B.L. Peptidoglycan recognition proteins involved in 1,3-β-D-glucan-dependent prophenoloxidase activation system of insect. J. Biol. Chem. 2004, 279, 3218–3227. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R. Peptidoglycan recognition proteins (PGRPs). Mol. Immunol. 2004, 40, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.W.; Soderhall, K. The prophenoloxidase activating system and associated proteins in invertebrates. Prog. Mol. Subcell. Biol. 1996, 15, 46–66. [Google Scholar] [PubMed]
- Tsuchiya, M.; Asahi, N.; Suzuoki, F.; Ashida, M.; Matsuura, S. Detection of peptidoglycan and β-glucan with silkworm larvae plasma test. FEMS Immunol. Med. Microbiol. 1996, 15, 129–134. [Google Scholar] [CrossRef]
- Yanaki, T.; Ito, W.; Tabata, K.; Kojima, T.; Norisuye, T.; Takano, N.; Fujita, H. Correlation between the antitumor activity of a polysaccharide schizophyllan and its triple-helical conformation in dilute aqueous solution. Biophys. Chem. 1983, 17, 337–342. [Google Scholar] [CrossRef]
- Ohno, N.; Emori, Y.; Yadomae, T.; Saito, K.; Masuda, A.; Oikawa, S. Reactivity of Limulus amoebocyte lysate towards (1→3)-β-D-glucans. Carbohydr. Res. 1990, 207, 311–318. [Google Scholar] [CrossRef]
- Young, S.H.; Dong, W.J.; Jacobs, R.R. Observation of a partially opened triple-helix conformation in 1→3-β-glucan by fluorescence resonance energy transfer spectroscopy. J. Biol. Chem. 2000, 275, 11874–11879. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Xu, X. Morphologies and conformation transition of lentinan in aqueous NaOH solution. Biopolymers 2004, 75, 187–195. [Google Scholar] [CrossRef]
- Rao, X.J.; Zhan, M.Y.; Pan, Y.M.; Liu, S.; Yang, P.J.; Yang, L.L.; Yu, X.Q. Immune functions of insect βGRPs and their potential application. Dev. Comp. Immunol. 2018, 83, 80–88. [Google Scholar] [CrossRef]
- Tanaka, S.; Aketagawa, J.; Takahashi, S.; Shibata, Y.; Tsumuraya, Y.; Hashimoto, Y. Inhibition of high-molecular-weight-(1→3)-β-D-glucan-dependent activation of a limulus coagulation factor G by laminaran oligosaccharides and curdlan degradation products. Carbohydr. Res. 1993, 244, 115–127. [Google Scholar] [CrossRef]
- Kanagawa, M.; Satoh, T.; Ikeda, A.; Adachi, Y.; Ohno, N.; Yamaguchi, Y. Structural insights into recognition of triple-helical β-glucans by an insect fungal receptor. J. Biol. Chem. 2011, 286, 29158–29165. [Google Scholar] [CrossRef]
- Takahasi, K.; Ochiai, M.; Horiuchi, M.; Kumeta, H.; Ogura, K.; Ashida, M.; Inagaki, F. Solution structure of the silkworm βGRP/GNBP3 N-terminal domain reveals the mechanism for β-1,3-glucan-specific recognition. Proc Natl Acad Sci. U.S.A. 2009, 106, 11679–11684. [Google Scholar] [CrossRef] [PubMed]
- Ohno, N.; Adachi, Y.; Ohsawa, M.; Sato, K.; Oikawa, S.; Yadomae, T. Conformational changes of the two different conformers of grifolan in sodium hydroxide, urea or dimethylsulfoxide solution. Chem. Pharm. Bull. 1987, 35, 2108–2113. [Google Scholar] [CrossRef] [PubMed]
- Miura, N.N.; Ohno, N.; Adachi, Y.; Yadomae, T. Strucural specificity of silkworm larvae plasma to (1→3)-β-D-glucans. Pharm. Pharmacol. Lett. 1996, 6, 111–114. [Google Scholar]
- Saito, H.; Yoshioka, Y.; Uehara, N.; Aketagawa, J.; Tanaka, S.; Shibata, Y. Relationship between conformation and biological response for (1→3)-β-D-glucans in the activation of coagulation factor G from limulus amebocyte lysate and host-mediated antitumor activity. Demonstration of single-helix conformation as a stimulant. Carbohydr. Res. 1991, 217, 181–190. [Google Scholar] [CrossRef]
- Aketagawa, J.; Tanaka, S.; Tamura, H.; Shibata, Y.; Saito, H. Activation of limulus coagulation factor G by several (1→3)-β-D-glucans: comparison of the potency of glucans with identical degree of polymerization but different conformations. J. Biochem. 1993, 113, 683–686. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.L.; Rice, P.J.; Graves, B.; Ensley, H.E.; Yu, H.; Brown, G.D.; Gordon, S.; Monteiro, M.A.; Papp-Szabo, E.; Lowman, D.W.; et al. Differential high-affinity interaction of dectin-1 with natural or synthetic glucans is dependent upon primary structure and is influenced by polymer chain length and side-chain branching. J. Pharmacol. Exp. Ther. 2008, 325, 115–123. [Google Scholar] [CrossRef]
- Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Beeman, R.W.; Brown, S.J.; Bucher, G.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar]
- Zhang, R.; Cho, H.Y.; Kim, H.S.; Ma, Y.G.; Osaki, T.; Kawabata, S.; Soderhall, K.; Lee, B.L. Characterization and properties of a 1,3-β-D-glucan pattern recognition protein of Tenebrio molitor larvae that is specifically degraded by serine protease during prophenoloxidase activation. J. Biol. Chem. 2003, 278, 42072–42079. [Google Scholar] [CrossRef] [PubMed]
- Nagi, N.; Ohno, N.; Adachi, Y.; Aketagawa, J.; Tamura, H.; Shibata, Y.; Tanaka, S.; Yadomae, T. Application of limulus test (G pathway) for the detection of different conformers of (1→3)-β-D-glucans. Biol. Pharm. Bull. 1993, 16, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Ishii, T.; Ikeda, Y.; Hoshino, A.; Tamura, H.; Aketagawa, J.; Tanaka, S.; Ohno, N. Characterization of β-glucan recognition site on C-type lectin, dectin 1. Infect. Immun. 2004, 72, 4159–4171. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, M.; Ashida, M. A pattern-recognition protein for β-1,3-glucan. The binding domain and the cDNA cloning of β-1,3-glucan recognition protein from the silkworm, Bombyx mori. J. Biol. Chem. 2000, 275, 4995–5002. [Google Scholar] [CrossRef] [PubMed]
- Tada, R.; Adachi, Y.; Ishibashi, K.; Tsubaki, K.; Ohno, N. Binding capacity of a barley β-D-glucan to the β-glucan recognition molecule dectin-1. J. Agric. Food Chem. 2008, 56, 1442–1450. [Google Scholar] [CrossRef] [PubMed]







| β-Glucan | BGRP | Conc. (nM) | KD (µM) | Ka (1/Ms) | Ka Error | Kd (1/s) | Kd Error | Rmax | R Equilibrium |
|---|---|---|---|---|---|---|---|---|---|
| SPG | Bm | 5650 | 0.29 | 2.09 × 105 | 1.26 × 104 | 6.16 × 10−2 | 6.16 × 10−2 | 0.2626 | 0.2496 |
| Tc | 5747 | 1.77 | 7.28 × 104 | 1.42 × 104 | 1.29 × 10−1 | 1.29 × 10−3 | 0.09497 | 0.07264 | |
| AT-SPG | Bm | 5650 | 0.20 | 3.40 × 105 | 3.59 × 104 | 6.79 × 10−2 | 6.79 × 10−2 | 0.07618 | 0.07358 |
| Tc | 5747 | 0.71 | 6.63 × 104 | 4.97 × 103 | 4.71 × 10−2 | 4.71 × 10−2 | 0.1853 | 0.1649 |
| BGRP CRDs | Primers | Sequence |
|---|---|---|
| Bm | Forward | 5′-CCAGATCTTACGAGGCACCACCGGCCAC-3′ |
| Reverse | 5′-GCGGATCCGAATTCTACTCCTGGTGTTAT-3′ | |
| Pi | Forward | 5′-GAGGATCCCAGCCGCGTGCGCAGCAGTAC-3′ |
| Reverse | 5′-GACCTGCAGCCCTCGAGACTCGTGTCAGCCGG-3′ | |
| Tc | Forward | 5′-GCGGATCCGAGTTTGAAGTTCCGGATGCT-3′ |
| Reverse | 5′-GACCTCGAGCTAGACTTTTCTTTGTCTAGTAA-3′ | |
| Tm | Forward | 5′-GCCAGATCTTTTGAGGTGCCAGATGCTTTG-3′ |
| Reverse | 5′-GCCGGATCCGTGTTCTTGCGGTGGAGCCTT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adachi, Y.; Ishii, M.; Kanno, T.; Tetsui, J.; Ishibashi, K.-i.; Yamanaka, D.; Miura, N.; Ohno, N. N-Terminal (1→3)-β-d-Glucan Recognition Proteins from Insects Recognize the Difference in Ultra-Structures of (1→3)-β-d-Glucan. Int. J. Mol. Sci. 2019, 20, 3498. https://doi.org/10.3390/ijms20143498
Adachi Y, Ishii M, Kanno T, Tetsui J, Ishibashi K-i, Yamanaka D, Miura N, Ohno N. N-Terminal (1→3)-β-d-Glucan Recognition Proteins from Insects Recognize the Difference in Ultra-Structures of (1→3)-β-d-Glucan. International Journal of Molecular Sciences. 2019; 20(14):3498. https://doi.org/10.3390/ijms20143498
Chicago/Turabian StyleAdachi, Yoshiyuki, Masaki Ishii, Takashi Kanno, Junko Tetsui, Ken-ichi Ishibashi, Daisuke Yamanaka, Noriko Miura, and Naohito Ohno. 2019. "N-Terminal (1→3)-β-d-Glucan Recognition Proteins from Insects Recognize the Difference in Ultra-Structures of (1→3)-β-d-Glucan" International Journal of Molecular Sciences 20, no. 14: 3498. https://doi.org/10.3390/ijms20143498
APA StyleAdachi, Y., Ishii, M., Kanno, T., Tetsui, J., Ishibashi, K.-i., Yamanaka, D., Miura, N., & Ohno, N. (2019). N-Terminal (1→3)-β-d-Glucan Recognition Proteins from Insects Recognize the Difference in Ultra-Structures of (1→3)-β-d-Glucan. International Journal of Molecular Sciences, 20(14), 3498. https://doi.org/10.3390/ijms20143498