Propolis Reduces the Expression of Autophagy-Related Proteins in Chondrocytes under Interleukin-1β Stimulus
Abstract
:1. Introduction
2. Results
2.1. Characterization of Polyphenols Present in the EEP
2.2. Cell Viability Analysis after Treatments
2.3. Effect of OA Induction on Autophagy-Related Proteins
2.4. Effect of EEP Treatment on Autophagy Protein in OA Chondrocytes
2.5. Effect of EEP Treatment on Cartilage Markers Expression in OA Chondrocytes
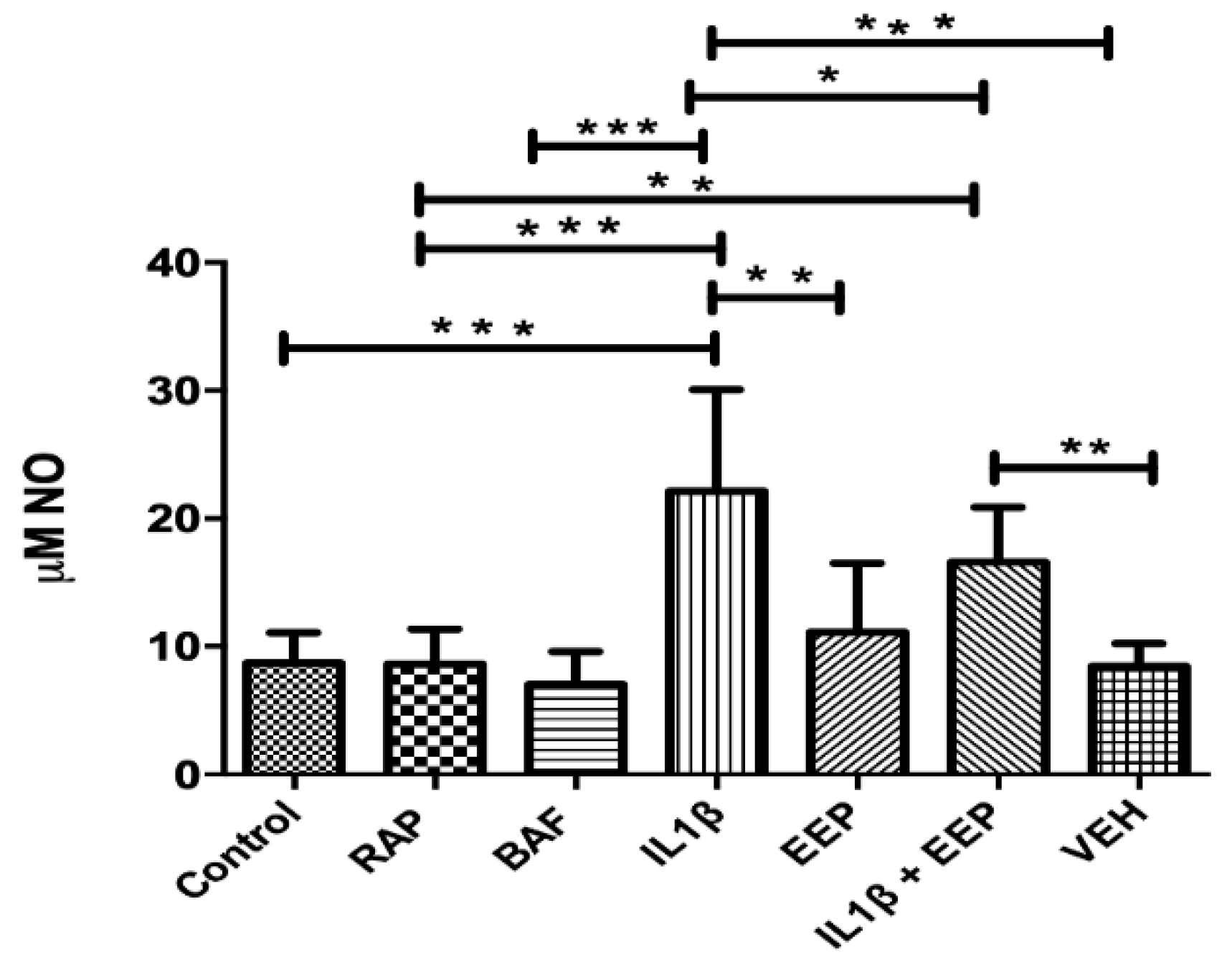
2.6. Effect of EEP Treatment on Chondrocytes Nitric Oxide Production Induced by the Inflammatory Stimulus
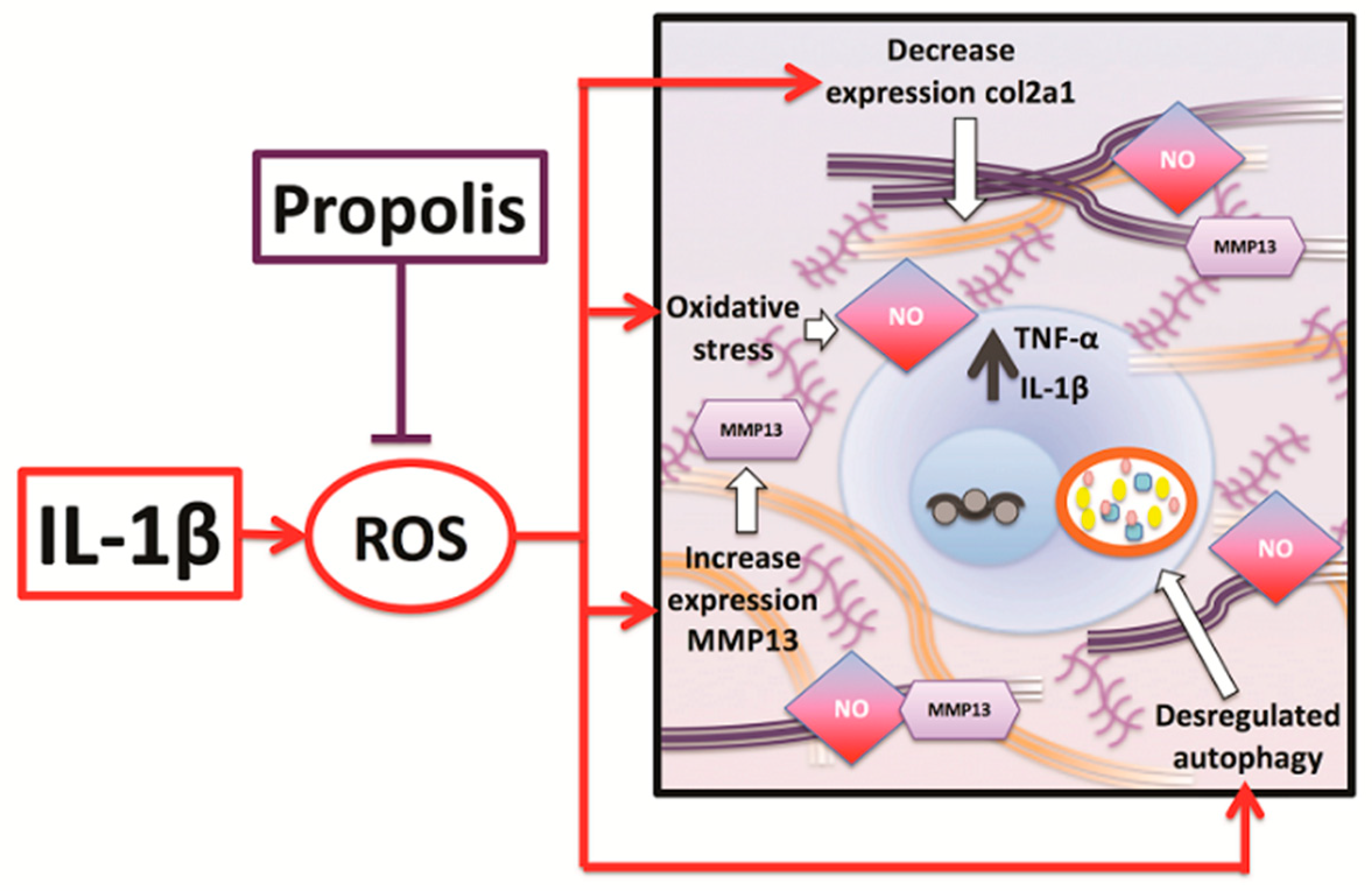
3. Discussion
4. Materials and Methods
4.1. Preparation and Characterization of Ethanolic Extract of Propolis
4.2. Cartilage Samples Collection and Primary Chondrocyte Culture
4.3. Cell Viability Analysis in Response to EEP Treatment
4.4. In Vitro Model of OA and Treatments
4.5. Western Blot Analysis
4.6. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
4.7. Measurement of NO Release
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AKT | Serine/Threonine Kinase |
| ATG | Autophagy-related proteins |
| ATG5 | Autophagy related 5 |
| BAF | Bafilomycin |
| BECN1 | Beclin1 |
| Col2a1 | Collagen type II alpha 1 chain |
| ECM | Extracellular matrix |
| EEP | Ethanolic extract of propolis |
| HPLC | High-performance liquid chromatography |
| HRP | Horseradish peroxidase |
| iNOS | Inducible nitric oxide synthase |
| IL-1β | interleukin 1 beta |
| LC3 | Microtubule associated protein 1 light chain 3 alpha (MAP1LC3A) |
| mRNA | Messenger RNA |
| MMPs | Matrix metalloproteinases |
| MMP-13 | Matrix metalloproteinase 13 |
| mTOR | Mammalian target of rapamycin |
| mTORC-1 | Mammalian target of rapamycin complex 1 |
| NO | Nitric oxide |
| OA | Osteoarthritis |
| OARSI | Osteoarthritis Research Society International |
| PB | Pinocembrin |
| RAP | Rapamycin |
| RNOS | Reactive Nitrogen and Oxygen Species |
| TNF-α | Tumor necrosis factor α |
| ULK1 | unc-51 like autophagy activating kinase 1 |
References
- Chen, D.; Shen, J.; Zhao, W.; Wang, T.; Han, L.; Hamilton, J.L.; Im, H.J. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 2017, 5, 16044. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, H.; Huang, L.; Welch, I.; Norley, C.; Holdsworth, D.W.; Beier, F.; Cai, D. Early Changes of Articular Cartilage and Subchondral Bone in The DMM Mouse Model of Osteoarthritis. Sci. Rep. 2018, 8, 2855. [Google Scholar] [CrossRef] [PubMed]
- Legendre, F.; Ollitrault, D.; Hervieu, M.; Bauge, C.; Maneix, L.; Goux, D.; Chajra, H.; Mallein-Gerin, F.; Boumediene, K.; Galera, P.; et al. Enhanced hyaline cartilage matrix synthesis in collagen sponge scaffolds by using siRNA to stabilize chondrocytes phenotype cultured with bone morphogenetic protein-2 under hypoxia. Tissue Eng. Part C Methods 2013, 19, 550–567. [Google Scholar] [CrossRef]
- Loeser, R.F. Aging processes and the development of osteoarthritis. Curr. Opin. Rheumatol. 2013, 25, 108–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobasheri, A.; Kalamegam, G.; Musumeci, G.; Batt, M.E. Chondrocyte and mesenchymal stem cell-based therapies for cartilage repair in osteoarthritis and related orthopaedic conditions. Maturitas 2014, 78, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Adamo, S.; Cetrullo, S.; Minguzzi, M.; Silvestri, Y.; Borzi, R.M.; Flamigni, F. MicroRNAs and Autophagy: Fine Players in the Control of Chondrocyte Homeostatic Activities in Osteoarthritis. Oxidative Med. Cell. Longev. 2017, 2017, 3720128. [Google Scholar] [CrossRef] [PubMed]
- Martel-Pelletier, J.; Barr, A.J.; Cicuttini, F.M.; Conaghan, P.G.; Cooper, C.; Goldring, M.B.; Goldring, S.R.; Jones, G.; Teichtahl, A.J.; Pelletier, J.P. Osteoarthritis. Nat. Rev. Dis. Primers 2016, 2, 16072. [Google Scholar] [CrossRef] [Green Version]
- Weng, T.; Xie, Y.; Yi, L.; Huang, J.; Luo, F.; Du, X.; Chen, L.; Liu, C.; Chen, D.; Chen, L. Loss of Vhl in cartilage accelerated the progression of age-associated and surgically induced murine osteoarthritis. Osteoarthr. Cartil. 2014, 22, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Schminke, B.; Miosge, N. Cartilage repair in vivo: The role of migratory progenitor cells. Curr. Rheumatol. Rep. 2014, 16, 461. [Google Scholar] [CrossRef]
- Todd Allen, R.; Robertson, C.M.; Harwood, F.L.; Sasho, T.; Williams, S.K.; Pomerleau, A.C.; Amiel, D. Characterization of mature vs aged rabbit articular cartilage: Analysis of cell density, apoptosis-related gene expression and mechanisms controlling chondrocyte apoptosis. Osteoarthr. Cartil. 2004, 12, 917–923. [Google Scholar] [CrossRef]
- Li, Y.; Wei, X.; Zhou, J.; Wei, L. The age-related changes in cartilage and osteoarthritis. BioMed Res. Int. 2013, 2013, 916530. [Google Scholar] [CrossRef]
- Hui, W.; Young, D.A.; Rowan, A.D.; Xu, X.; Cawston, T.E.; Proctor, C.J. Oxidative changes and signalling pathways are pivotal in initiating age-related changes in articular cartilage. Ann. Rheum. Dis. 2016, 75, 449–458. [Google Scholar] [CrossRef]
- Mobasheri, A.; Matta, C.; Zakany, R.; Musumeci, G. Chondrosenescence: Definition, hallmarks and potential role in the pathogenesis of osteoarthritis. Maturitas 2015, 80, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Lei, M.; Wang, J.G.; Xiao, D.M.; Fan, M.; Wang, D.P.; Xiong, J.Y.; Chen, Y.; Ding, Y.; Liu, S.L. Resveratrol inhibits interleukin 1beta-mediated inducible nitric oxide synthase expression in articular chondrocytes by activating SIRT1 and thereby suppressing nuclear factor-kappaB activity. Eur. J. Pharmacol. 2012, 674, 73–79. [Google Scholar] [CrossRef]
- Johnson, C.I.; Argyle, D.J.; Clements, D.N. In vitro models for the study of osteoarthritis. Vet. J. 2016, 209, 40–49. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Cai, G.Q.; Peng, J.P.; Chen, X.D. Autophagy protects chondrocytes from glucocorticoids-induced apoptosis via ROS/Akt/FOXO3 signaling. Osteoarthr. Cartil. 2015, 23, 2279–2287. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.L.; Smith, B.J.; Lo, D.F.; Chyu, M.C.; Dunn, D.M.; Chen, C.H.; Kwun, I.S. Dietary polyphenols and mechanisms of osteoarthritis. J. Nutr. Biochem. 2012, 23, 1367–1377. [Google Scholar] [CrossRef]
- Oliveira, M.Z.; Albano, M.B.; Stirma, G.A.; Namba, M.M.; Vidigal, L.; Cunha, L. Intra-articular viscosupplementation of hyaluronic acids in an experimental osteoarthritis model. Rev. Bras. Ortop. 2018, 53, 293–299. [Google Scholar] [CrossRef]
- Ding, Q.H.; Ye, C.Y.; Chen, E.M.; Zhang, W.; Wang, X.H. Emodin ameliorates cartilage degradation in osteoarthritis by inhibiting NF-kappaB and Wnt/beta-catenin signaling in-vitro and in-vivo. Int. Immunopharmacol. 2018, 61, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Terman, A.; Kurz, T.; Navratil, M.; Arriaga, E.A.; Brunk, U.T. Mitochondrial turnover and aging of long-lived postmitotic cells: The mitochondrial-lysosomal axis theory of aging. Antioxid. Redox Signal. 2010, 12, 503–535. [Google Scholar] [CrossRef]
- Lotz, M.K.; Carames, B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nat. Rev. Rheumatol. 2011, 7, 579–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tashiro, K.; Shishido, M.; Fujimoto, K.; Hirota, Y.; Yo, K.; Gomi, T.; Tanaka, Y. Age-related disruption of autophagy in dermal fibroblasts modulates extracellular matrix components. Biochem. Biophys. Res. Commun. 2014, 443, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Linton, P.J.; Gurney, M.; Sengstock, D.; Mentzer, R.M., Jr.; Gottlieb, R.A. This old heart: Cardiac aging and autophagy. J. Mol. Cell. Cardiol. 2015, 83, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, K.Y.; Park, S.G.; Yu, S.N.; Kim, Y.W.; Nam, H.W.; An, H.H.; Kim, Y.W.; Ahn, S.C. Mitochondrial ROS activates ERK/autophagy pathway as a protected mechanism against deoxypodophyllotoxin-induced apoptosis. Oncotarget 2017, 8, 111581–111596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carames, B.; Kiosses, W.B.; Akasaki, Y.; Brinson, D.C.; Eap, W.; Koziol, J.; Lotz, M.K. Glucosamine activates autophagy in vitro and in vivo. Arthritis Rheum. 2013, 65, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Fullgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Reviews. Mol. Cell Biol. 2014, 15, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and cellular immune responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Carames, B.; Olmer, M.; Kiosses, W.B.; Lotz, M.K. The relationship of autophagy defects to cartilage damage during joint aging in a mouse model. Arthritis Rheumatol. 2015, 67, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.D.; Wu, D.; Gu, J.H.; Ge, J.B.; Wu, J.C.; Han, R.; Liang, Z.Q.; Qin, Z.H. The pro-survival role of autophagy depends on Bcl-2 under nutrition stress conditions. PLoS ONE 2013, 8, e63232. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: A double-edged sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Lockshin, R.A.; Zakeri, Z. Apoptosis, autophagy, and more. Int. J. Biochem. Cell Biol. 2004, 36, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Muriach, M.; Flores-Bellver, M.; Romero, F.J.; Barcia, J.M. Diabetes and the brain: Oxidative stress, inflammation, and autophagy. Oxidative Med. Cell. Longev. 2014, 2014, 102158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Kouroku, Y.; Fujita, E.; Tanida, I.; Ueno, T.; Isoai, A.; Kumagai, H.; Ogawa, S.; Kaufman, R.J.; Kominami, E.; Momoi, T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007, 14, 230–239. [Google Scholar] [CrossRef]
- Arroyo, D.S.; Gaviglio, E.A.; Peralta Ramos, J.M.; Bussi, C.; Rodriguez-Galan, M.C.; Iribarren, P. Autophagy in inflammation, infection, neurodegeneration and cancer. Int. Immunopharmacol. 2014, 18, 55–65. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Markaki, M.; Palikaras, K.; Tavernarakis, N. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 2013, 1833, 3448–3459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieland, H.A.; Michaelis, M.; Kirschbaum, B.J.; Rudolphi, K.A. Osteoarthritis-an untreatable disease? Nat. Rev. Drug Discov. 2005, 4, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Serafini, M.; Maiani, G.; Ferro-Luzzi, A. Alcohol-free red wine enhances plasma antioxidant capacity in humans. J. Nutr. 1998, 128, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Reinli, K.; Block, G. Phytoestrogen content of foods-a compendium of literature values. Nutr. Cancer 1996, 26, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.D.; Yuan, R.Y.; Wu, Q.; Li, S.S.; Shao, S.; Xu, Y.J.; Hao, X.H.; Wang, L.S. Assessment of flavonoids and volatile compounds in tea infusions of water lily flowers and their antioxidant activities. Food Chem. 2015, 187, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Manzano, S.; Santos-Buelga, C.; Perez-Alonso, J.J.; Rivas-Gonzalo, J.C.; Escribano-Bailon, M.T. Characterization of the mean degree of polymerization of proanthocyanidins in red wines using liquid chromatography-mass spectrometry (LC-MS). J. Agric. Food Chem. 2006, 54, 4326–4332. [Google Scholar] [CrossRef]
- Pan, M.H.; Lai, C.S.; Wu, J.C.; Ho, C.T. Epigenetic and disease targets by polyphenols. Curr. Pharm. Des. 2013, 19, 6156–6185. [Google Scholar] [CrossRef]
- Shukla, S.; Meeran, S.M.; Katiyar, S.K. Epigenetic regulation by selected dietary phytochemicals in cancer chemoprevention. Cancer Lett. 2014, 355, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Ayissi, V.B.; Ebrahimi, A.; Schluesenner, H. Epigenetic effects of natural polyphenols: A focus on SIRT1-mediated mechanisms. Mol. Nutr. Food Res. 2014, 58, 22–32. [Google Scholar] [CrossRef]
- Schiano, C.; Vietri, M.T.; Grimaldi, V.; Picascia, A.; Pascale, M.R.; Napoli, C. Epigenetic-related therapeutic challenges in cardiovascular disease. Trends Pharmacol. Sci. 2015, 36, 226–235. [Google Scholar] [CrossRef]
- Joven, J.; Micol, V.; Segura-Carretero, A.; Alonso-Villaverde, C.; Menendez, J.A.; Bioactive Food Components Platform. Polyphenols and the modulation of gene expression pathways: Can we eat our way out of the danger of chronic disease? Crit. Rev. Food Sci. Nutr. 2014, 54, 985–1001. [Google Scholar] [CrossRef]
- Bag, A.; Bag, N. Tea Polyphenols and Prevention of Epigenetic Aberrations in Cancer. J. Nat. Sci. Biol. Med. 2018, 9, 2–5. [Google Scholar] [CrossRef]
- Abdul, Q.A.; Yu, B.P.; Chung, H.Y.; Jung, H.A.; Choi, J.S. Epigenetic modifications of gene expression by lifestyle and environment. Arch. Pharmacal Res. 2017, 40, 1219–1237. [Google Scholar] [CrossRef]
- Pallauf, K.; Rimbach, G. Autophagy, polyphenols and healthy ageing. Ageing Res. Rev. 2013, 12, 237–252. [Google Scholar] [CrossRef]
- Portal-Nunez, S.; Esbrit, P.; Alcaraz, M.J.; Largo, R. Oxidative stress, autophagy, epigenetic changes and regulation by miRNAs as potential therapeutic targets in osteoarthritis. Biochem. Pharmacol. 2016, 108, 1–10. [Google Scholar] [CrossRef]
- Sabe, A.A.; Elmadhun, N.Y.; Dalal, R.S.; Robich, M.P.; Sellke, F.W. Resveratrol regulates autophagy signaling in chronically ischemic myocardium. J. Thorac. Cardiovasc. Surg. 2014, 147, 792–798. [Google Scholar] [CrossRef]
- Poulose, S.M.; Fisher, D.R.; Bielinski, D.F.; Gomes, S.M.; Rimando, A.M.; Schauss, A.G.; Shukitt-Hale, B. Restoration of stressor-induced calcium dysregulation and autophagy inhibition by polyphenol-rich acai (Euterpe spp.) fruit pulp extracts in rodent brain cells in vitro. Nutrition 2014, 30, 853–862. [Google Scholar] [CrossRef]
- Pietrocola, F.; Marino, G.; Lissa, D.; Vacchelli, E.; Malik, S.A.; Niso-Santano, M.; Zamzami, N.; Galluzzi, L.; Maiuri, M.C.; Kroemer, G. Pro-autophagic polyphenols reduce the acetylation of cytoplasmic proteins. Cell Cycle 2012, 11, 3851–3860. [Google Scholar] [CrossRef] [Green Version]
- Shabalala, S.; Muller, C.J.F.; Louw, J.; Johnson, R. Polyphenols, autophagy and doxorubicin-induced cardiotoxicity. Life Sci. 2017, 180, 160–170. [Google Scholar] [CrossRef]
- Budisan, L.; Gulei, D.; Jurj, A.; Braicu, C.; Zanoaga, O.; Cojocneanu, R.; Pop, L.; Raduly, L.; Barbat, A.; Moldovan, A.; et al. Inhibitory Effect of CAPE and Kaempferol in Colon Cancer Cell Lines-Possible Implications in New Therapeutic Strategies. Int. J. Mol. Sci. 2019, 20, 1199. [Google Scholar] [CrossRef]
- Volpert, R.; Elstner, E.F. Biochemical activities of propolis extracts. II. Photodynamic activities. Z. Fur Nat. C 1993, 48, 858–862. [Google Scholar] [CrossRef]
- Cardile, V.; Panico, A.; Gentile, B.; Borrelli, F.; Russo, A. Effect of propolis on human cartilage and chondrocytes. Life Sci. 2003, 73, 1027–1035. [Google Scholar] [CrossRef]
- El-Ghazaly, M.A.; El-Naby, D.H.; Khayyal, M.T. The influence of irradiation on the potential chondroprotective effect of aqueous extract of propolis in rats. Int. J. Radiat. Biol. 2011, 87, 254–262. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.Z.; Song, J.K.; Sun, J.L.; Li, Y.J.; Zhou, S.B.; Zhang, T.T.; Du, G.H. Pinocembrin protects human brain microvascular endothelial cells against fibrillar amyloid-beta(1–40) injury by suppressing the MAPK/NF-kappaB inflammatory pathways. BioMed Res. Int. 2014, 2014, 470393. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, W.; Li, L.; Wu, S.; Du, G. Pinocembrin protects the brain against ischemia-reperfusion injury and reverses the autophagy dysfunction in the penumbra area. Molecules 2014, 19, 15786–15798. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, B.; Xiong, C.; Yue, Z. Pinocembrin inhibits matrix metalloproteinase expression in chondrocytes. IUBMB Life 2015, 67, 36–41. [Google Scholar] [CrossRef]
- Tao, J.; Shen, C.; Sun, Y.; Chen, W.; Yan, G. Neuroprotective effects of pinocembrin on ischemia/reperfusion-induced brain injury by inhibiting autophagy. Biomed. Pharmacother. 2018, 106, 1003–1010. [Google Scholar] [CrossRef]
- Jeon, H.; Im, G.I. Autophagy in osteoarthritis. Connect. Tissue Res. 2017, 58, 497–508. [Google Scholar] [CrossRef]
- Almonte-Becerril, M.; Navarro-Garcia, F.; Gonzalez-Robles, A.; Vega-Lopez, M.A.; Lavalle, C.; Kouri, J.B. Cell death of chondrocytes is a combination between apoptosis and autophagy during the pathogenesis of Osteoarthritis within an experimental model. Apoptosis 2010, 15, 631–638. [Google Scholar] [CrossRef]
- Musumeci, G.; Castrogiovanni, P.; Trovato, F.M.; Weinberg, A.M.; Al-Wasiyah, M.K.; Alqahtani, M.H.; Mobasheri, A. Biomarkers of Chondrocyte Apoptosis and Autophagy in Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 20560–20575. [Google Scholar] [CrossRef] [Green Version]
- Pasupuleti, V.R.; Sammugam, L.; Ramesh, N.; Gan, S.H. Honey, Propolis, and Royal Jelly: A Comprehensive Review of Their Biological Actions and Health Benefits. Oxidative Med. Cell. Longev. 2017, 2017, 1259510. [Google Scholar] [CrossRef]
- Burdock, G.A. Review of the biological properties and toxicity of bee propolis (propolis). Food Chem. Toxicol. 1998, 36, 347–363. [Google Scholar] [CrossRef]
- Zabaiou, N.; Fouache, A.; Trousson, A.; Baron, S.; Zellagui, A.; Lahouel, M.; Lobaccaro, J.A. Biological properties of propolis extracts: Something new from an ancient product. Chem. Phys. Lipids 2017, 207, 214–222. [Google Scholar] [CrossRef]
- Cuevas, A.; Saavedra, N.; Cavalcante, M.F.; Salazar, L.A.; Abdalla, D.S. Identification of microRNAs involved in the modulation of pro-angiogenic factors in atherosclerosis by a polyphenol-rich extract from propolis. Arch. Biochem. Biophys. 2014, 557, 28–35. [Google Scholar] [CrossRef]
- Veloz, J.J.; Alvear, M.; Salazar, L.A. Antimicrobial and Antibiofilm Activity against Streptococcus mutans of Individual and Mixtures of the Main Polyphenolic Compounds Found in Chilean Propolis. BioMed Res. Int. 2019, 2019, 7602343. [Google Scholar] [CrossRef]
- Veloz, J.J.; Saavedra, N.; Lillo, A.; Alvear, M.; Barrientos, L.; Salazar, L.A. Antibiofilm Activity of Chilean Propolis on Streptococcus mutans Is Influenced by the Year of Collection. BioMed Res. Int. 2015, 2015, 291351. [Google Scholar] [CrossRef]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef]
- Wu, T.T.; Li, W.M.; Yao, Y.M. Interactions between Autophagy and Inhibitory Cytokines. Int. J. Biol. Sci. 2016, 12, 884–897. [Google Scholar] [CrossRef]
- Qian, M.; Fang, X.; Wang, X. Autophagy and inflammation. Clin. Transl. Med. 2017, 6, 24. [Google Scholar] [CrossRef]
- Jia, G.; Cheng, G.; Gangahar, D.M.; Agrawal, D.K. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol. Cell Biol. 2006, 84, 448–454. [Google Scholar] [CrossRef]
- Gu, Y.T.; Chen, J.; Meng, Z.L.; Ge, W.Y.; Bian, Y.Y.; Cheng, S.W.; Xing, C.K.; Yao, J.L.; Fu, J.; Peng, L. Research progress on osteoarthritis treatment mechanisms. Biomed. Pharmacother. 2017, 93, 1246–1252. [Google Scholar] [CrossRef]
- Henrotin, Y.; Kurz, B.; Aigner, T. Oxygen and reactive oxygen species in cartilage degradation: Friends or foes? Osteoarthr. Cartil. 2005, 13, 643–654. [Google Scholar] [CrossRef]
- Xuan, H.; Yuan, W.; Chang, H.; Liu, M.; Hu, F. Anti-inflammatory effects of Chinese propolis in lipopolysaccharide-stimulated human umbilical vein endothelial cells by suppressing autophagy and MAPK/NF-kappaB signaling pathway. Inflammopharmacology 2019, 27, 561–571. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Li, L.H.; Rao, Y.K.; Ju, T.C.; Nai, Y.S.; Chen, Y.W.; Hua, K.F. Mechanistic insight into the attenuation of gouty inflammation by Taiwanese green propolis via inhibition of the NLRP3 inflammasome. J. Cell. Physiol. 2019, 234, 4081–4094. [Google Scholar] [CrossRef]
- Lee, E.J.; Kang, M.K.; Kim, Y.H.; Kim, D.Y.; Oh, H.; Kim, S.I.; Oh, S.Y.; Kang, Y.H. Dietary Chrysin Suppresses Formation of Actin Cytoskeleton and Focal Adhesion in AGE-Exposed Mesangial Cells and Diabetic Kidney: Role of Autophagy. Nutrients 2019, 11, 127. [Google Scholar] [CrossRef]
- Goutas, A.; Syrrou, C.; Papathanasiou, I.; Tsezou, A.; Trachana, V. The autophagic response to oxidative stress in osteoarthritic chondrocytes is deregulated. Free Radic. Biol. Med. 2018, 126, 122–132. [Google Scholar] [CrossRef]
- Maroudas, A.; Palla, G.; Gilav, E. Racemization of aspartic acid in human articular cartilage. Connect. Tissue Res. 1992, 28, 161–169. [Google Scholar] [CrossRef]
- Tiku, M.L.; Madhan, B. Preserving the longevity of long-lived type II collagen and its implication for cartilage therapeutics. Ageing Res. Rev. 2016, 28, 62–71. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Martinet, W.; De Meyer, G.R.; Andries, L.; Herman, A.G.; Kockx, M.M. In situ detection of starvation-induced autophagy. J. Histochem. Cytochem. 2006, 54, 85–96. [Google Scholar] [CrossRef]
- Ahn, M.R.; Kunimasa, K.; Kumazawa, S.; Nakayama, T.; Kaji, K.; Uto, Y.; Hori, H.; Nagasawa, H.; Ohta, T. Correlation between antiangiogenic activity and antioxidant activity of various components from propolis. Mol. Nutr. Food Res. 2009, 53, 643–651. [Google Scholar] [CrossRef]
- Kang, S.W.; Yoo, S.P.; Kim, B.S. Effect of chondrocyte passage number on histological aspects of tissue-engineered cartilage. Bio-Med Mater. Eng. 2007, 17, 269–276. [Google Scholar]
- Tennant, J.R. Evaluation of the Trypan Blue Technique for Determination of Cell Viability. Transplantation 1964, 2, 685–694. [Google Scholar] [CrossRef]
- Faruqi, T.R.; Erzurum, S.C.; Kaneko, F.T.; DiCorleto, P.E. Role of nitric oxide in poly(I-C)-induced endothelial cell expression of leukocyte adhesion molecules. Am. J. Physiol. 1997, 273, H2490–H2497. [Google Scholar] [CrossRef]





| Nº | Rt (Min) | λmax (nm) | [M-H]− (m/z) | Ion Formula | Error (ppm) | Compound |
|---|---|---|---|---|---|---|
| 1 | 7.3 | 292, 323 | 179.0353 | C9H7O4 | 1.6 | Caffeic acid |
| 2 | 11.4 | 310 | 163.0399 | C9H7O3 | −2.2 | p-Coumaric acid |
| 3 | 12.6 | 298, 320 | 193.0508 | C10H9O4 | 0.9 | Isoferulic acid |
| 4 | 14.3 | 295, 322 | 193.0506 | C10H9O4 | 0.0 | Ferulic acid |
| 5 | 16.8 | 292 | 287.0565 | C15H11O6 | 1.3 | Dihydrokaempferol |
| 6 | 18.0 | 230 | 121.0281 | C7H5O2 | −6.1 | Benzoic acid |
| 7 | 22.4 | 295, 320 | 207.0658 | C11H11O4 | 0.3 | 3,4-Dimethyl-caffeic acid |
| 8 | 30.4 | 295, 322 | 207.0664 | C11H11O4 | 0.0 | 3,4-Dimethyl-caffeic acid isomer |
| 9 | 31.2 | 285 | 285.0768 | C16H13O5 | −0.2 | Pinobanksin-5-methyl ether |
| 10 | 32.3 | 308 | 177.0559 | C10H9O3 | 1.0 | p-Coumaric acid methyl ester |
| 11 | 33.3 | 255, 369 | 301.0351 | C15H9O7 | −0.8 | Quercetin |
| 12 | 34.3 | 291 | 271.0619 | C15H11O5 | 2.5 | Pinobanksin |
| 13 | 37.6 | 252, 350 | 285.0404 | C15H9O6 | −0.1 | Luteolin |
| 14 | 39.5 | 256, 357 | 315.0504 | C16H11O7 | −1.9 | Quercetin-3-methyl ether |
| 15 | 44.0 | 265, 366 | 285.0410 | C15H9O6 | 1.8 | Kaempferol |
| 16 | 44.6 | 286 | 269.0826 | C16H13O4 | 2.7 | Pinocembrin-5-methyl ether |
| 17 | 46.3 | 267, 338 | 269.0448 | C15H9O5 | 2.6 | Apigenin |
| 18 | 46.9 | 254, 370 | 315.0510 | C16H11O7 | −0.1 | Isorhamnetin |
| 19 | 49.7 | 266, 352 | 299.0549 | C16H11O6 | −4.0 | Kaempferol-methyl ether |
| 20 | 50.5 | 311 | 173.0621 | C11H9O2 | 7.6 | Chrysin-5-methyl ether |
| 21 | 50.8 | 265, 310 | 267.0663 | C16H11O4 | −3.2 | Cinnamylidenacetic acid |
| 22 | 51.2 | 290 | 329.0674 | C17H13O7 | 2.1 | Unknown |
| 23 | 53.6 | 296 | 313.0714 | C17H13O6 | 1.1 | Pinobanksin acetate derivative |
| 24 | 55.3 | 255, 369 | 315.0508 | C16H11O7 | −0.7 | Rhamnetin |
| 25 | 57.1 | 290 | 255.0664 | C15H11O4 | 0.5 | Pinocembrin |
| 26 | 59.4 | 327 | 269.0818 | C16H13O4 | 0.4 | Caffeic acid benzyl ester |
| 27 | 59.8 | 300, 325 | 247.0977 | C14H15O4 | 0.7 | Caffeic acid isoprenyl ester (isomer) |
| 28 | 60.3 | 290 | 285.0766 | C16H13O5 | −0.8 | Unknown |
| 29 | 61.1 | 292 | 313.0727 | C17H13O6 | 3.1 | Pinobanksin-3-O-acetate |
| 30 | 61.4 | 300, 325 | 247.0988 | C14H15O4 | 4.9 | Caffeic acid isoprenyl ester (isomer) |
| 31 | 62.0 | 301, 325 | 247.0986 | C14H15O4 | 3.9 | Caffeic acid isoprenyl ester (isomer) |
| 32 | 63.3 | 268, 314 | 253.0515 | C15H9O4 | 3.6 | Chrysin |
| 33 | 65.7 | 298, 325 | 283.0987 | C17H15O4 | 4.0 | Caffeic acid phenylethyl ester (CAPE) |
| 34 | 66.5 | 265, 359 | 269.0467 | C15H9O5 | 4.5 | Galangin |
| 35 | 69.3 | 268, 332 | 283.0624 | C16H11O5 | 4.2 | Acacetin |
| 36 | 69.8 | 311 | 253.0883 | C16H13O3 | 5.0 | p-Coumaric acid benzyl ester |
| 37 | 70.4 | 298, 325 | 283.0987 | C17H15O4 | 3.9 | Caffeic acid derivative (phenylethyl ester) |
| 38 | 70.9 | 298, 323 | 261.1138 | C15H17O4 | 2.3 | Caffeic acid derivative (hexenoate ester) |
| 39 | 72.2 | 298, 324 | 261.1145 | C15H17O4 | 4.9 | Caffeic acid derivative (hexenoate ester) |
| 40 | 72.7 | 290 | 327.0904 | C18H15O6 | 9.0 | Pinobanksin-3-O-propionate |
| 41 | 74.1 | 247, 295, 325 | 295.0993 | C18H15O4 | 5.7 | Caffeic acid cinnamyl ester |
| 42 | 75.7 | 298 | 267.1033 | C17H15O3 | 2.3 | Unknown |
| 43 | 76.2 | 300, 327 | 269.0825 | C16H13O4 | 2.0 | Caffeic acid derivative |
| 44 | 78.7 | 289 | 433.1313 | C25H21O7 | 4.7 | Pinocembrin methoxyphenylpropionate derivative |
| 45 | 82.4 | nd | 417.1376 | C25H21O6 | 7.9 | Pinobanksin phenylpropionate derivative |
| 46 | 82.9 | 286 | 353.1062 | C20H17O6 | 9.0 | Pinobanksin-3-O-pentenoate |
| 47 | 83.3 | 293 | 341.1048 | C19H17O6 | 8.2 | Pinobanksin-3-O-butyrate/isobutyrate |
| 48 | 88.3 | nd | 311.2249 | C18H31O4 | 6.8 | Unknown |
| 49 | 92.5 | 292 | 355.1220 | C20H19O6 | 9.3 | Pinobanksin-3-O-pentanoate/2-methylbutyrate |
| 50 | 96.1 | 291 | 403.1238 | C24H19O6 | 12.6 | Pinobanksin-3-O-phenylpropionate |
| 51 | 97.7 | 291 | 369.1397 | C21H21O6 | 14.4 | Pinobanksin-3-O-hexanoate |
| 52 | 99.6 | 283 | 293.2148 | C18H29O3 | 8.7 | Unknown |
| 53 | 103.6 | 283 | 281.2509 | C18H33O2 | 9.5 | Unknown |
| Gene Symbol | Sequence Forward (5′-3′) | Sequence Reverse (5′-3′) |
|---|---|---|
| COL2A1 | GGT GAC TAC TGG ATA GAC CCC AAC CAA | TGA AGT GGA AGC CGC CAT TGA TG |
| MMP13 | GAA TTA AGG AGC ATG GCG AC | TAA GGA GTG GCC GAA CTC AT |
| ATG5 | CGT CCT GTG GCT GCA GAT G | AAG GAC ACA CTT CTT TGA GGA GAT C |
| MAPLC3A/B (LC3I/II) | GCC TTC TTC CTG CTG GTG AAC | AGC CGT CCT CGT CTT TCT CC |
| AKT 1 | ATG GCA CCT TCA TTG GCT AC | CCC AGC AGC TTC AGG TAC TC |
| ACTB | AGA CCA CCT TCA ACT CGA TCA T | ACT CGT CAT ACT CCT GCT TGC T |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arias, C.; Saavedra, N.; Saavedra, K.; Alvear, M.; Cuevas, A.; Stuchi Maria-Engler, S.; Abdalla, D.S.P.; Salazar, L.A. Propolis Reduces the Expression of Autophagy-Related Proteins in Chondrocytes under Interleukin-1β Stimulus. Int. J. Mol. Sci. 2019, 20, 3768. https://doi.org/10.3390/ijms20153768
Arias C, Saavedra N, Saavedra K, Alvear M, Cuevas A, Stuchi Maria-Engler S, Abdalla DSP, Salazar LA. Propolis Reduces the Expression of Autophagy-Related Proteins in Chondrocytes under Interleukin-1β Stimulus. International Journal of Molecular Sciences. 2019; 20(15):3768. https://doi.org/10.3390/ijms20153768
Chicago/Turabian StyleArias, Consuelo, Nicolás Saavedra, Kathleen Saavedra, Marysol Alvear, Alejandro Cuevas, Silvya Stuchi Maria-Engler, Dulcineia S. P. Abdalla, and Luis A. Salazar. 2019. "Propolis Reduces the Expression of Autophagy-Related Proteins in Chondrocytes under Interleukin-1β Stimulus" International Journal of Molecular Sciences 20, no. 15: 3768. https://doi.org/10.3390/ijms20153768
APA StyleArias, C., Saavedra, N., Saavedra, K., Alvear, M., Cuevas, A., Stuchi Maria-Engler, S., Abdalla, D. S. P., & Salazar, L. A. (2019). Propolis Reduces the Expression of Autophagy-Related Proteins in Chondrocytes under Interleukin-1β Stimulus. International Journal of Molecular Sciences, 20(15), 3768. https://doi.org/10.3390/ijms20153768







