Peroxisome Maintenance Depends on De Novo Peroxisome Formation in Yeast Mutants Defective in Peroxisome Fission and Inheritance
Abstract
:1. Introduction
2. Results
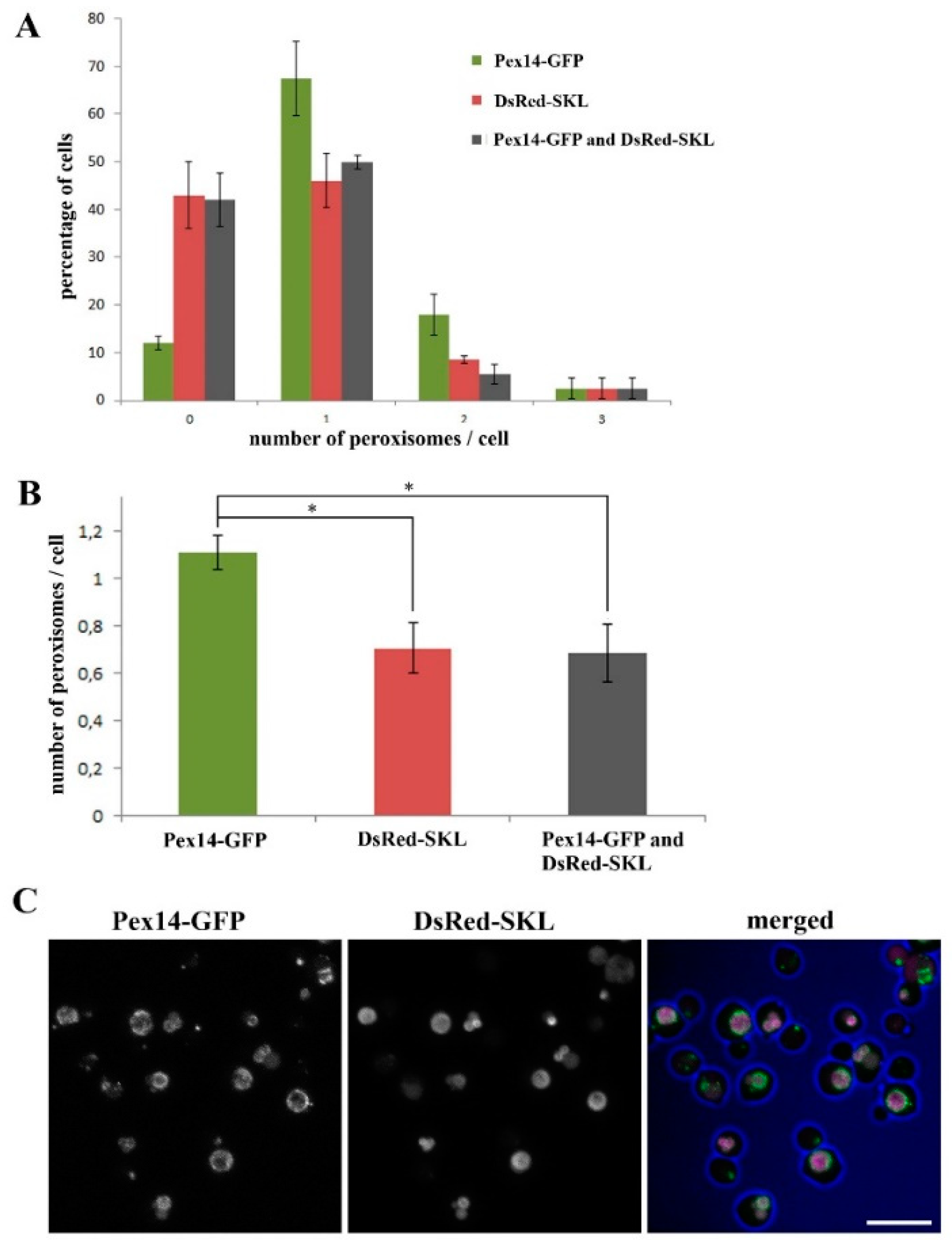
2.1. Almost All H. polymorpha pex11 Cells Contain Peroxisomes
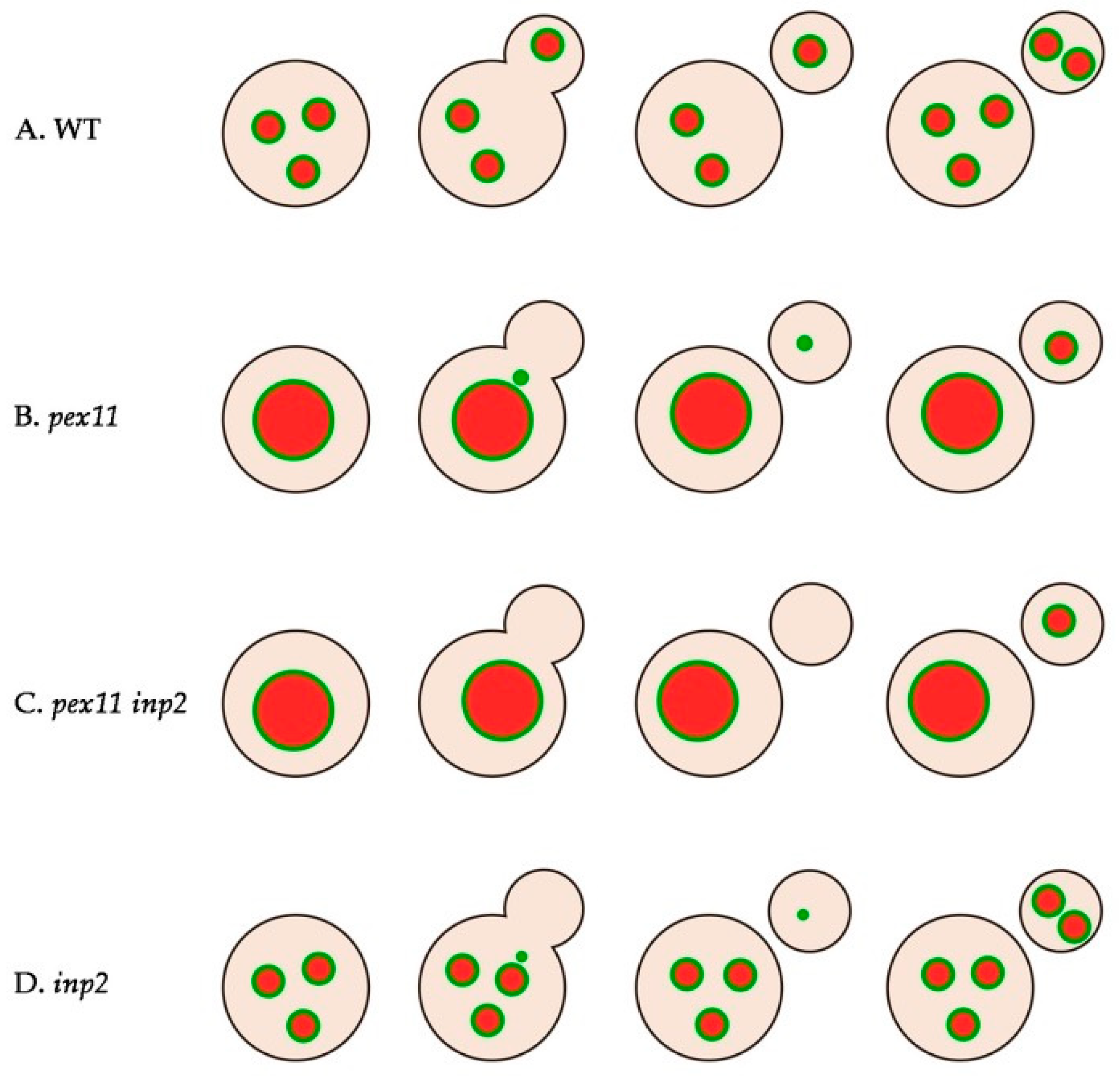
2.2. Peroxisomes can be Inherited to H. polymorpha pex11 Daughter Cells
2.3. Peroxisome Inheritance is not Detected in pex11 inp2 Cells
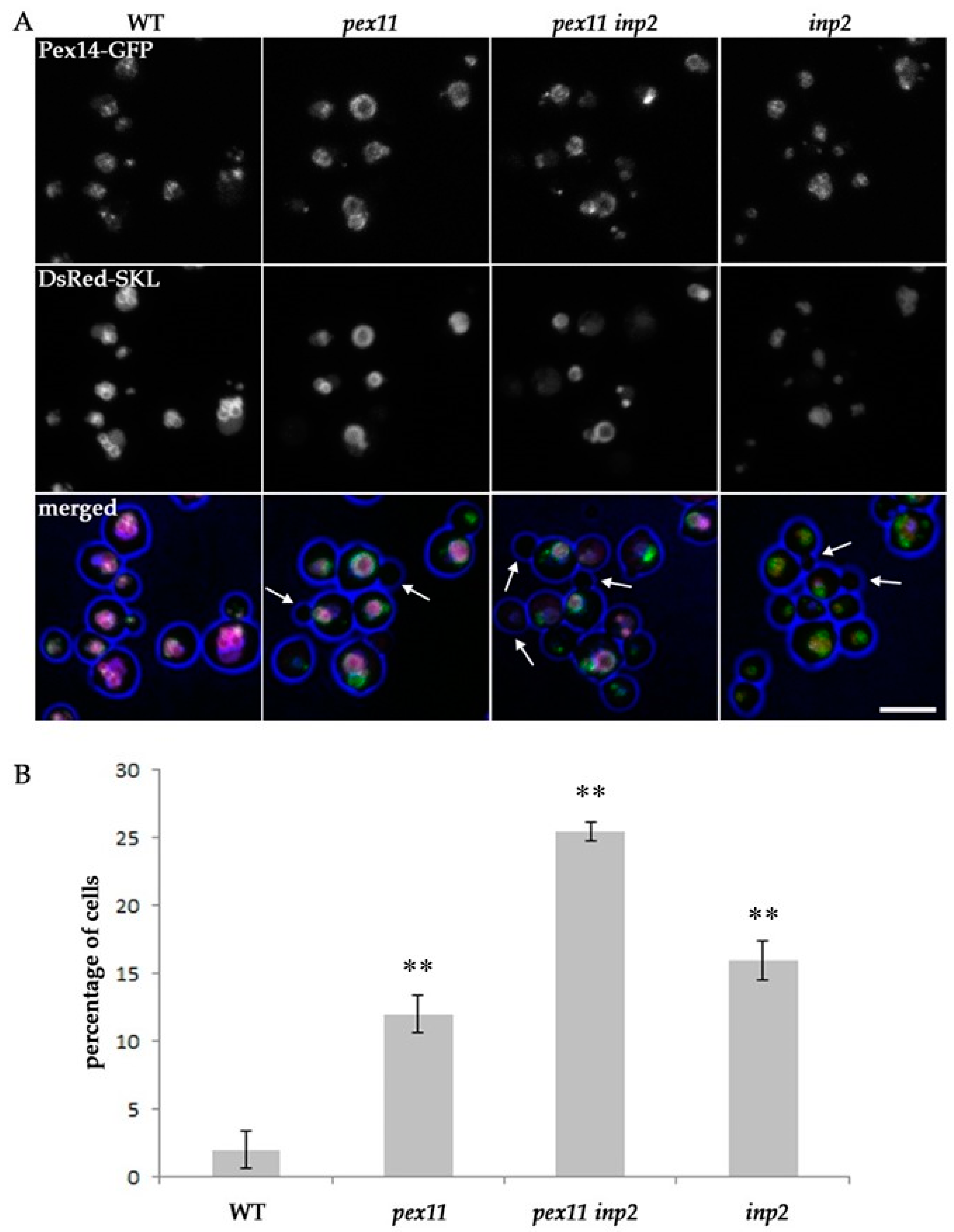
2.4. Block of Fission and Inheritance Increases the Number of Cells Devoid of Peroxisomes
2.5. Peroxisomes Reappear in Buds of pex11 inp2 Cells
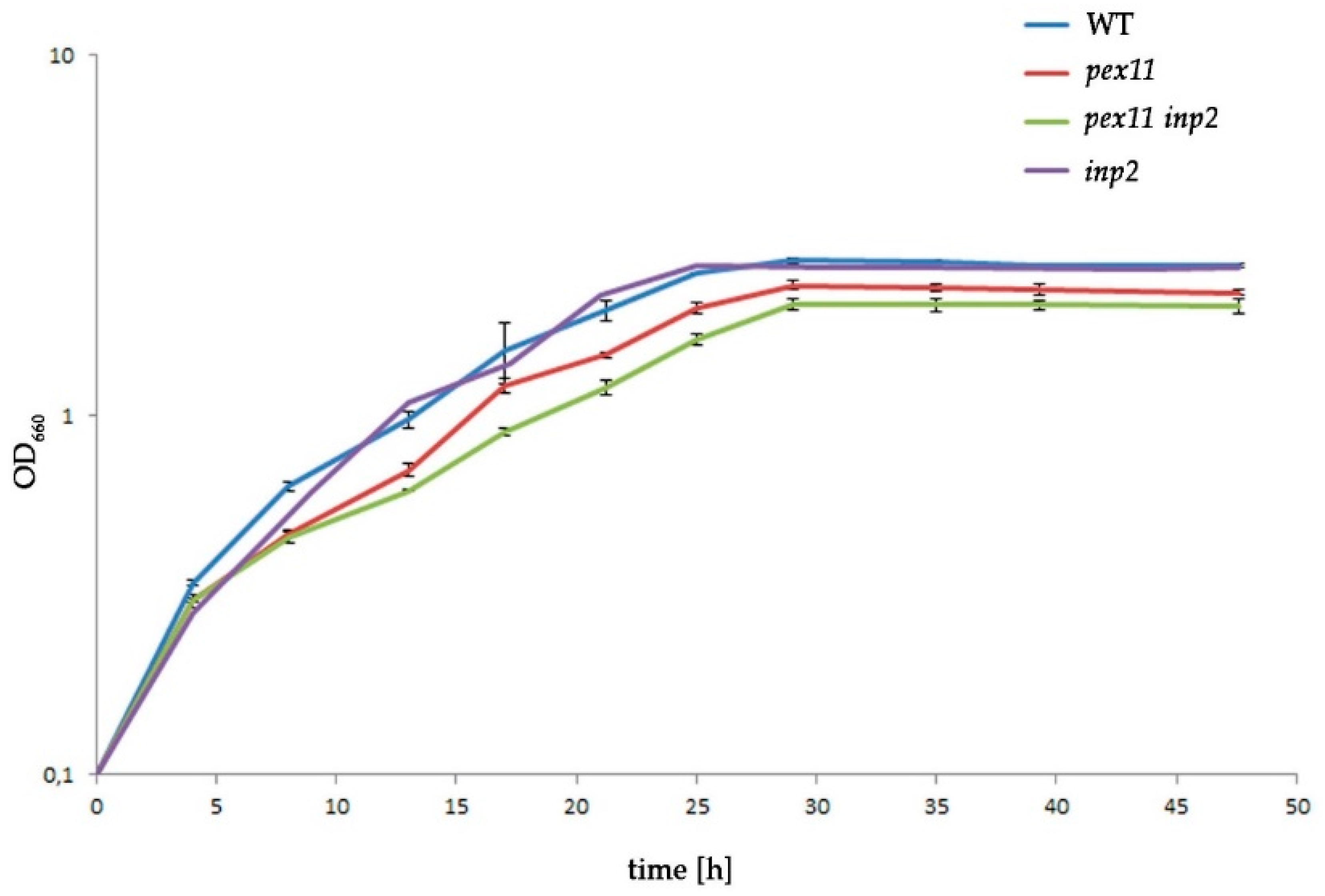
2.6. The Combined Fission and Inheritance Defects of H. polymorpha pex11 inp2 Cells Increase the Doubling Time on Methanol Medium
2.7. inp2 Cells are Not Fully Defective in Peroxisome Inheritance
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Molecular Techniques
4.3. Construction of Yeast Strains
4.4. Fluorescence Microscopy
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kohlwein, S.D.; Veenhuis, M.; van der Klei, I.J. Lipid droplets and peroxisomes: Key players in cellular lipid homeostasis or a matter of fat--store ’em up or burn ’em down. Genetics 2013, 193, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant Peroxisomes: Biogenesis and Function. Plant Cell 2012, 24, 2279–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanders, R.J.; Waterham, H.R. Peroxisomal disorders: The single peroxisomal enzyme deficiencies. Biochim. Biophys. Acta BBA Bioenerg. 2006, 1763, 1707–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veenhuis, M.; Harder, W. Microbodies in yeasts: Structure, function and biogenesis. Microbiol. Sci. 1988, 5, 347–351. [Google Scholar] [PubMed]
- Dixit, E.; Boulant, S.; Zhang, Y.; Lee, A.S.; Odendall, C.; Shum, B.; Hacohen, N.; Chen, Z.J.; Whelan, S.P.; Fransen, M.; et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 2010, 141, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Singh, R.; Williams, C.P.; Van Der Klei, I.J. Stress exposure results in increased peroxisomal levels of yeast Pnc1 and Gpd1, which are imported via a piggy-backing mechanism. Biochim. Biophys. Acta BBA Bioenerg. 2016, 1863, 148–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Klei, I.J.; Yurimoto, H.; Sakai, Y.; Veenhuis, M. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta BBA Bioenerg. 2006, 1763, 1453–1462. [Google Scholar] [CrossRef] [Green Version]
- Titorenko, V.I.; Smith, J.J.; Szilard, R.K.; Rachubinski, R.A. Peroxisome Biogenesis in the Yeast Yarrowia lipolytica. Cell Biophys. 2000, 32, 21–26. [Google Scholar] [CrossRef]
- Titorenko, V.I.; Mullen, R.T. Peroxisome biogenesis: The peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 2006, 174, 11–17. [Google Scholar] [CrossRef]
- Van Der Zand, A.; Braakman, I.; Tabak, H.F. Peroxisomal Membrane Proteins Insert into the Endoplasmic Reticulum. Mol. Biol. Cell 2010, 21, 2057–2065. [Google Scholar] [CrossRef] [Green Version]
- Van Der Zand, A.; Gent, J.; Braakman, I.; Tabak, H.F. Biochemically Distinct Vesicles from the Endoplasmic Reticulum Fuse to Form Peroxisomes. Cell 2012, 149, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, A.; Mattie, S.; Prudent, J.; McBride, H.M. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 2017, 542, 251–254. [Google Scholar] [CrossRef]
- Fujiki, Y.; Lazarow, P.B. Biogenesis of Peroxisomes. Annu. Rev. Cell Boil. 1985, 1, 489–530. [Google Scholar]
- Motley, A.M.; Ward, G.P.; Hettema, E.H. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 2008, 121, 1633–1640. [Google Scholar] [CrossRef] [Green Version]
- Nagotu, S.; Saraya, R.; Otzen, M.; Veenhuis, M.; Van Der Klei, I.J. Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim. Biophys. Acta BBA Bioenerg. 2008, 1783, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Fagarasanu, M.; Fagarasanu, A.; Tam, Y.Y.C.; Aitchison, J.D.; Rachubinski, R.A. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2005, 169, 765–775. [Google Scholar] [CrossRef]
- Krikken, A.M.; Veenhuis, M.; Van Der Klei, I.J. Hansenula polymorpha pex11 cells are affected in peroxisome retention. FEBS J. 2009, 276, 1429–1439. [Google Scholar] [CrossRef]
- Hoepfner, D.; Berg, M.V.D.; Philippsen, P.; Tabak, H.F.; Hettema, E.H. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2001, 155, 979–990. [Google Scholar] [CrossRef]
- Fagarasanu, A.; Fagarasanu, M.; Eitzen, G.A.; Aitchison, J.D.; Rachubinski, R.A. The Peroxisomal Membrane Protein Inp2p Is the Peroxisome-Specific Receptor for the Myosin V Motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 2006, 10, 587–600. [Google Scholar] [CrossRef]
- Saraya, R.; Cepińska, M.N.; Kiel, J.A.; Veenhuis, M.; Van Der Klei, I.J. A conserved function for Inp2 in peroxisome inheritance. Biochim. Biophys. Acta BBA Bioenerg. 2010, 1803, 617–622. [Google Scholar] [CrossRef] [Green Version]
- Lam, S.K.; Yoda, N.; Schekman, R. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2010, 107, 21523–21528. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.; Joshi, S.; Subramani, S. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2011, 108, 9113–9118. [Google Scholar] [CrossRef] [Green Version]
- Hoepfner, D.; Schildknegt, D.; Braakman, I.; Philippsen, P.; Tabak, H.F. Contribution of the Endoplasmic Reticulum to Peroxisome Formation. Cell 2005, 122, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Kragt, A.; Voorn-Brouwer, T.; Berg, M.V.D.; Distel, B. Endoplasmic Reticulum-directed Pex3p Routes to Peroxisomes and Restores Peroxisome Formation in a Saccharomyces cerevisiae pex3Δ Strain. J. Biol. Chem. 2005, 280, 34350–34357. [Google Scholar] [CrossRef]
- David, C.; Koch, J.; Oeljeklaus, S.; Laernsack, A.; Melchior, S.; Wiese, S.; Schummer, A.; Erdmann, R.; Warscheid, B.; Brocard, C. A Combined Approach of Quantitative Interaction Proteomics and Live-cell Imaging Reveals a Regulatory Role for Endoplasmic Reticulum (ER) Reticulon Homology Proteins in Peroxisome Biogenesis. Mol. Cell. Proteom. 2013, 12, 2408–2425. [Google Scholar] [CrossRef] [Green Version]
- Mast, F.D.; Jamakhandi, A.; Saleem, R.A.; Dilworth, D.J.; Rogers, R.S.; Rachubinski, R.A.; Aitchison, J.D. Peroxins Pex30 and Pex29 Dynamically Associate with Reticulons to Regulate Peroxisome Biogenesis from the Endoplasmic Reticulum. J. Biol. Chem. 2016, 291, 15408–15427. [Google Scholar] [CrossRef] [Green Version]
- Joshi, A.S.; Huang, X.; Choudhary, V.; Levine, T.P.; Hu, J.; Prinz, W.A. A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol. 2016, 215, 515–529. [Google Scholar] [CrossRef] [Green Version]
- Motley, A.M.; Hettema, E.H. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 2007, 178, 399–410. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.; Opalinski, L.; Landgraf, C.; Costello, J.; Schrader, M.; Krikken, A.M.; Knoops, K.; Kram, A.M.; Volkmer, R.; Van Der Klei, I.J. The membrane remodeling protein Pex11p activates the GTPase Dnm1p during peroxisomal fission. Proc. Natl. Acad. Sci. USA 2015, 112, 6377–6382. [Google Scholar] [CrossRef] [Green Version]
- Cepińska, M.N.; Veenhuis, M.; Van Der Klei, I.J.; Nagotu, S. Peroxisome Fission is Associated with Reorganization of Specific Membrane Proteins. Traffic 2011, 12, 925–937. [Google Scholar] [CrossRef]
- Kuravi, K.; Nagotu, S.; Krikken, A.M.; Sjollema, K.; Deckers, M.; Erdmann, R.; Veenhuis, M.; Van Der Klei, I.J. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 2006, 119, 3994–4001. [Google Scholar] [CrossRef]
- Huber, A.; Koch, J.; Kragler, F.; Brocard, C.; Hartig, A. A Subtle Interplay between Three Pex11 Proteins Shapes De Novo Formation and Fission of Peroxisomes. Traffic 2012, 13, 157–167. [Google Scholar] [CrossRef]
- Kim, P.K.; Mullen, R.T.; Schumann, U.; Lippincott-Schwartz, J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 2006, 173, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Waterham, H.R.; Zhang, K.; Sha, J.; Harter, M.L. Peroxisomal Targeting, Import, and Assembly of Alcohol Oxidase in Pichia pastoris. J. Cell Biol. 1997, 139, 1419–1431. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Mast, F.D.; Fagarasanu, A.; Rachubinski, D.A.; Eitzen, G.A.; Dacks, J.B.; Rachubinski, R.A. Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J. Cell Biol. 2009, 187, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Otzen, M.; Rucktäschel, R.; Emmrich, K.; Krikken, A.M.; Van Der Klei, I.J.; Thoms, S.; Erdmann, R.; Klei, I.J. Pex19p Contributes to Peroxisome Inheritance in the Association of Peroxisomes to Myo2p. Traffic 2012, 13, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Wróblewska, J.P.; Cruz-Zaragoza, L.D.; Yuan, W.; Schummer, A.; Chuartzman, S.G.; De Boer, R.; Oeljeklaus, S.; Schuldiner, M.; Zalckvar, E.; Warscheid, B.; et al. Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. Biochim. Biophys. Acta BBA Bioenerg. 2017, 1864, 1656–1667. [Google Scholar] [CrossRef]
- Mast, F.D.; Herricks, T.; Strehler, K.M.; Miller, L.R.; Saleem, R.A.; Rachubinski, R.A.; Aitchison, J.D. ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 2018, 217, 2087–2102. [Google Scholar] [CrossRef] [Green Version]
- Sudbery, E.; Gleeson, P.A.; Veale, M.A.; Ledeboer, A.M.; Zoetmulder, M.C. Hansenula polymorpha as a novel yeast system for the expression of heterologous genes. Biochem. Soc. Trans. 1988, 16, 1081–1083. [Google Scholar] [CrossRef]
- Yuan, W. Origin and Growth of Peroxisomes in Yeast. Ph.D. Thesis, University of Groningen, Groningen, The Netherlands, 2016. [Google Scholar]
- Opaliński, Ł.; Kiel, J.A.K.W.; Williams, C.; Veenhuis, M.; van der Klei, I.J. Membrane curvature during peroxisome fission requires Pex11. EMBO J. 2011, 30, 5–16. [Google Scholar] [CrossRef]
- Sambrook, J. Molecular Cloning: A Laboratory Manual; CSHL Press: Cold Spring Harbor, NY, USA, 2001; p. 1365. [Google Scholar]
- Faber, K.N.; Haima, P.; Harder, W.; Veenhuis, M.; Ab, G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 1994, 25, 305–310. [Google Scholar] [CrossRef]




| Strain | Description | Reference |
|---|---|---|
| NCYC 495, leu1.1, ura3 | Wild type | [39] |
| inp2 | INP2::URA3 | [20] |
| WT Pex14-GFP DsRed-SKL | pHIPZ-Pex14-GFP::ZEO; pHIPN4-DsRed-SKL::NAT | This study |
| pex11 DsRed-SKL | PEX11::HPH; pHIPN4-DsRed-SKL::NAT | [40] |
| pex11 Pex14-GFP DsRed-SKL | PEX11::HPH; HIPN4-DsRed-SKL::NAT; pHIPZ-Pex14-GFP::ZEO | This study |
| pex11 DsRed-SKL | PEX11::URA3; pHIPN4-DsRed-SKL::NAT | [41] |
| pex11 inp2 Pex14-GFP DsRed-SKL | PEX11::URA3; INP2::HPH; pHIPZ-Pex14-GFP::ZEO; pHIPN4-DsRed-SKL::NAT | This study |
| inp2 Pex14-GFP DsRed-SKL | INP2::URA3; pHIPZ-Pex14-GFP::ZEO; pHIPN4-DsRed-SKL::NAT | This study |
| Name | Description | Reference |
|---|---|---|
| pHIPZ-Pex14-GFP | pHIPZ plasmid containing the 3′-end of the PEX14 gene fused in frame to GFP; ZeoR; AmpR | [30] |
| pHIPN4_DsRed-SKL | pHIPN4 plasmid containing DsRed-SKL under the control of PAOX; NatR, AmpR | [30] |
| pARM001 | pHIPH plasmid containing gene encoding C-terminal part of Pex14 fused to mCherry; HphR, AmpR | [6] |
| Name | Sequence (5′-3′) |
|---|---|
| JWR156 | TTTTTATTTTATCATTTTCTATCCTCACGAGATCGCATCAAGGCACCGCTTAACCCACACACCATAGCTTCAA |
| JWR157 | TGATGTCGAGAATCAAAAACGCTGTTGCCAGCAGCGTCGCGAGCTTCAGGCGTTTTCGACACTGGATGGC |
| JWR158 | CGAACTGGTGGTTAAGAGCG |
| JWR159 | GCTTTTGGCTGCGGGAACGT |
| JWR031 | TCCTGCCAGAATTGAACTAG |
| JWR032 | GTACGGGTAATTAACGACAC |
| JWR160 | CACAATTGGAGCAGGACAAG |
| JWR161 | GCCGTCGTCCTTGAAGA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wróblewska, J.P.; van der Klei, I.J. Peroxisome Maintenance Depends on De Novo Peroxisome Formation in Yeast Mutants Defective in Peroxisome Fission and Inheritance. Int. J. Mol. Sci. 2019, 20, 4023. https://doi.org/10.3390/ijms20164023
Wróblewska JP, van der Klei IJ. Peroxisome Maintenance Depends on De Novo Peroxisome Formation in Yeast Mutants Defective in Peroxisome Fission and Inheritance. International Journal of Molecular Sciences. 2019; 20(16):4023. https://doi.org/10.3390/ijms20164023
Chicago/Turabian StyleWróblewska, Justyna P., and Ida J. van der Klei. 2019. "Peroxisome Maintenance Depends on De Novo Peroxisome Formation in Yeast Mutants Defective in Peroxisome Fission and Inheritance" International Journal of Molecular Sciences 20, no. 16: 4023. https://doi.org/10.3390/ijms20164023
APA StyleWróblewska, J. P., & van der Klei, I. J. (2019). Peroxisome Maintenance Depends on De Novo Peroxisome Formation in Yeast Mutants Defective in Peroxisome Fission and Inheritance. International Journal of Molecular Sciences, 20(16), 4023. https://doi.org/10.3390/ijms20164023





