Highly Efficient and Heritable Targeted Mutagenesis in Wheat via the Agrobacterium tumefaciens-Mediated CRISPR/Cas9 System
Abstract
:1. Introduction
2. Results
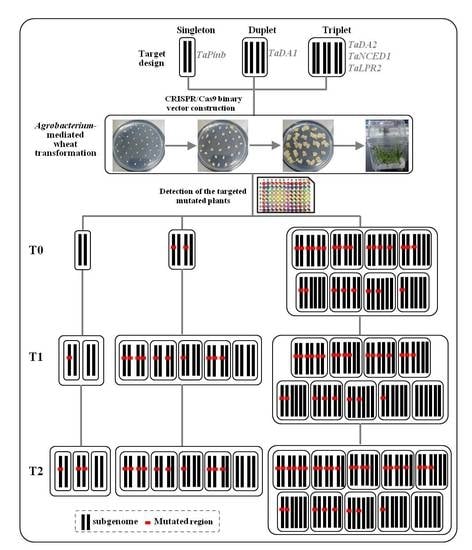
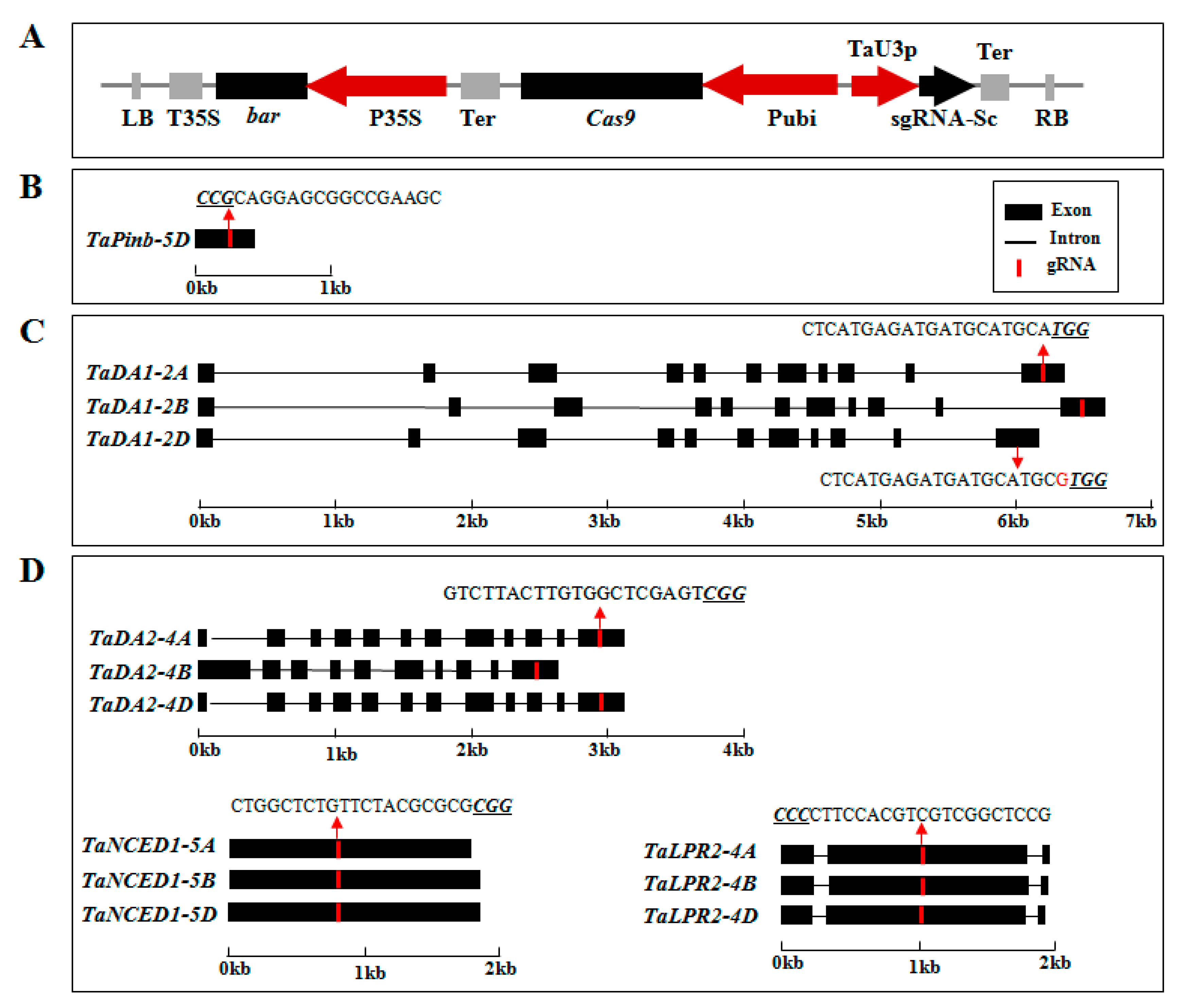
2.1. sgRNA Design and Vector Construction
2.2. CRISPR/Cas9 Induced Highly Efficient Mutagenesis in Four Genes but Not in TaPinb in T0 Plants
2.3. Inheritance and Stability of Targeted Mutations Induced by CRISPR/Cas9 in the T1 and T2 Generations
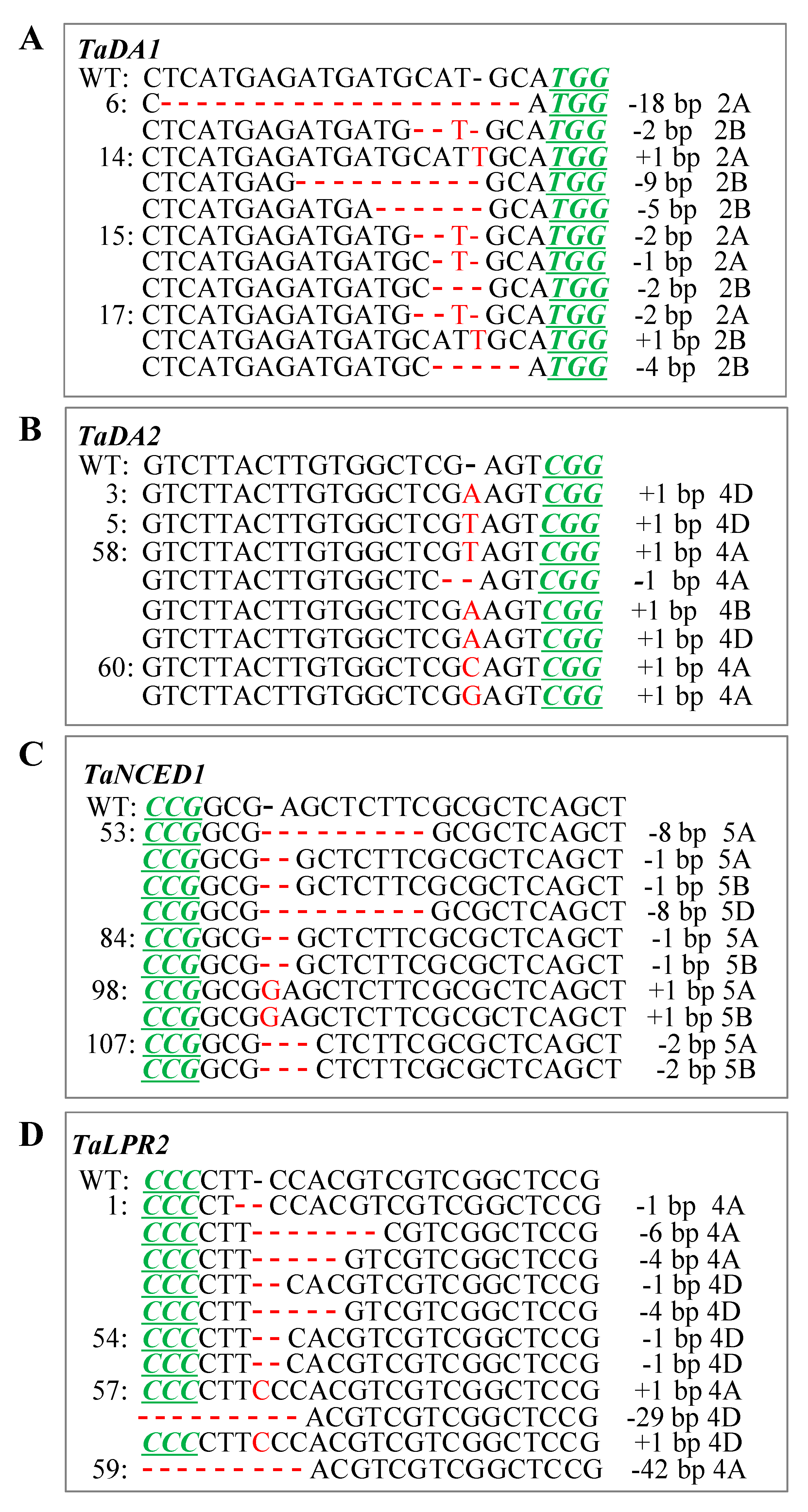
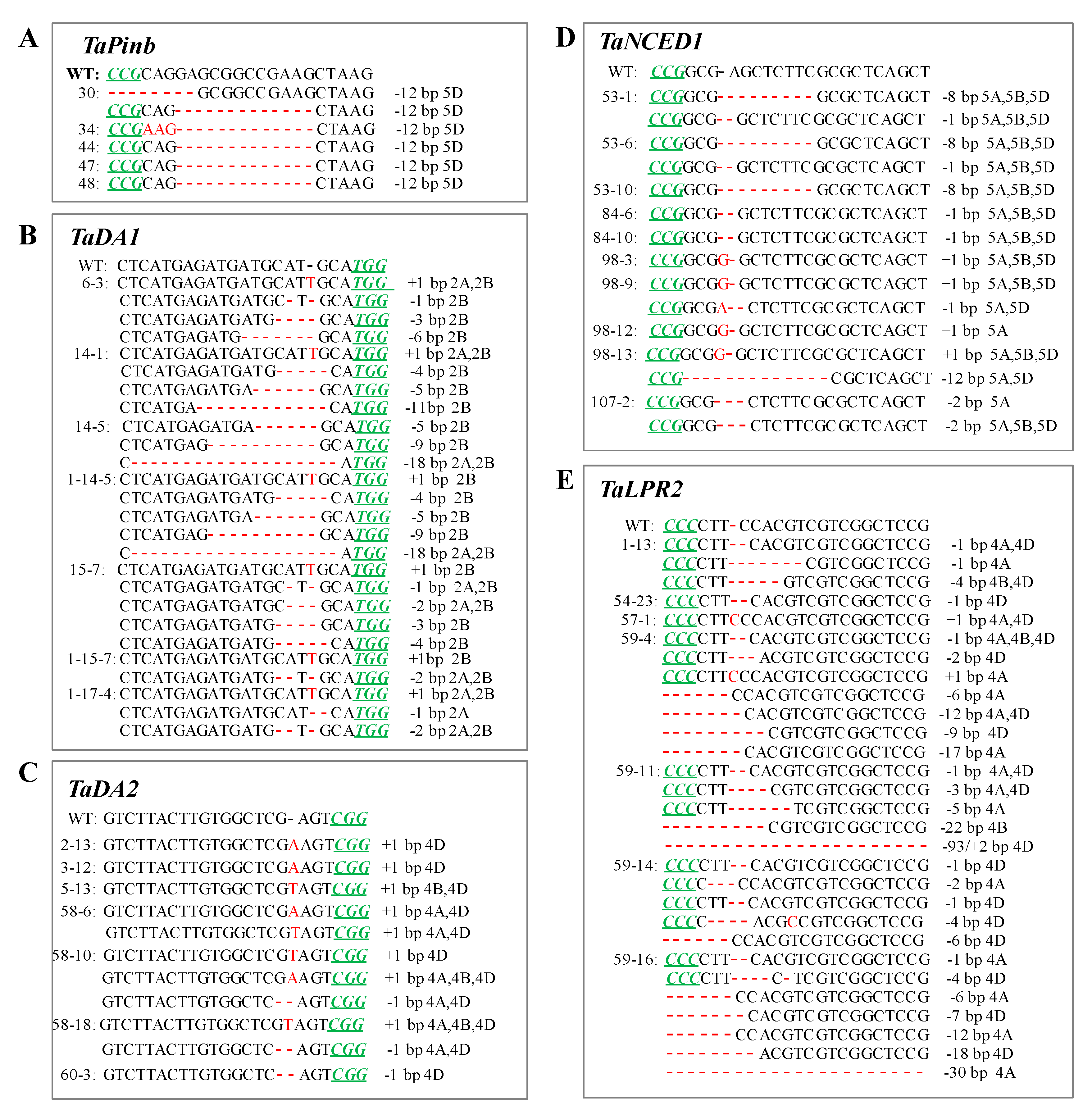
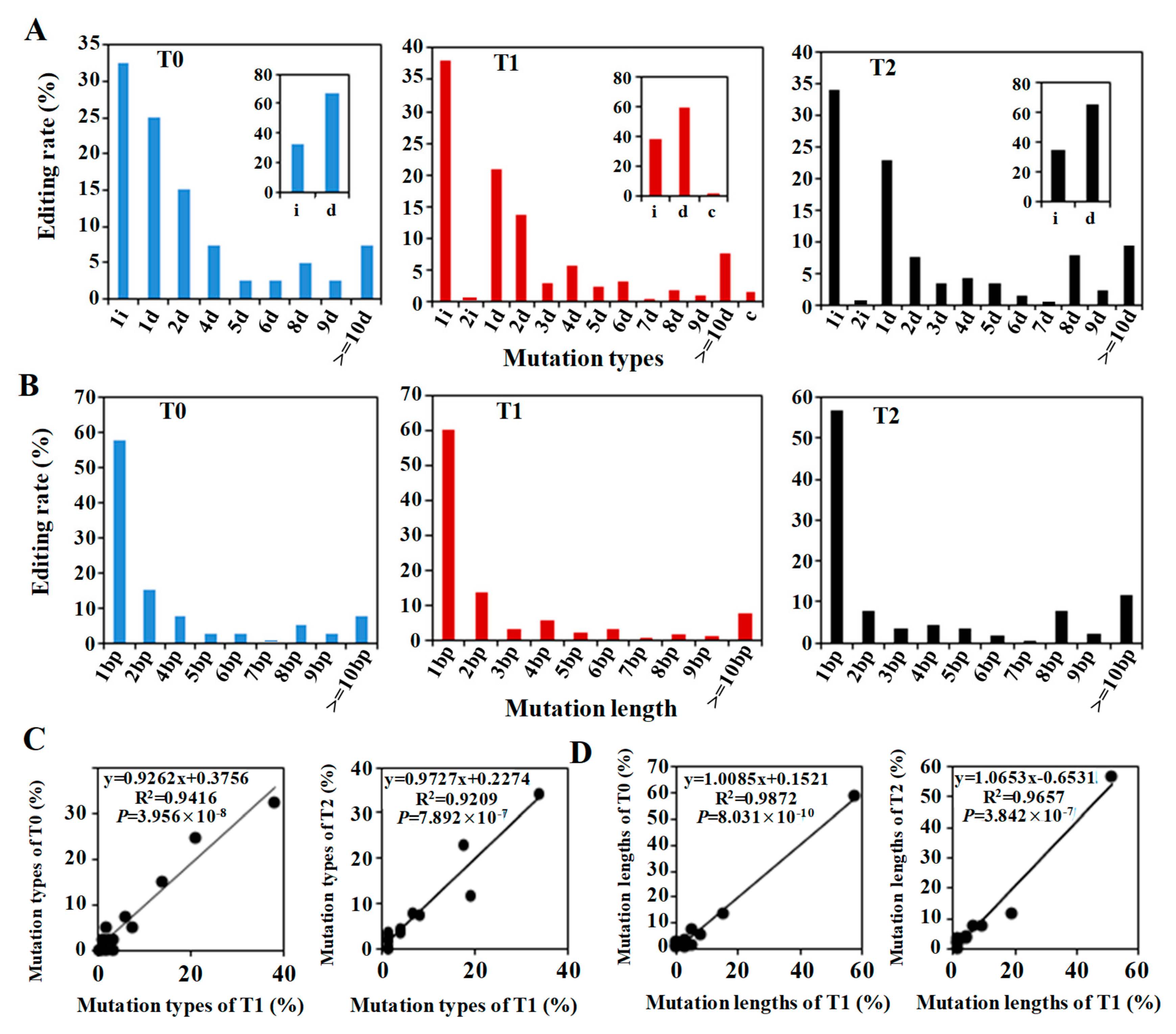
2.4. CRISPR/Cas9-Induced Mutation Types Across Generations
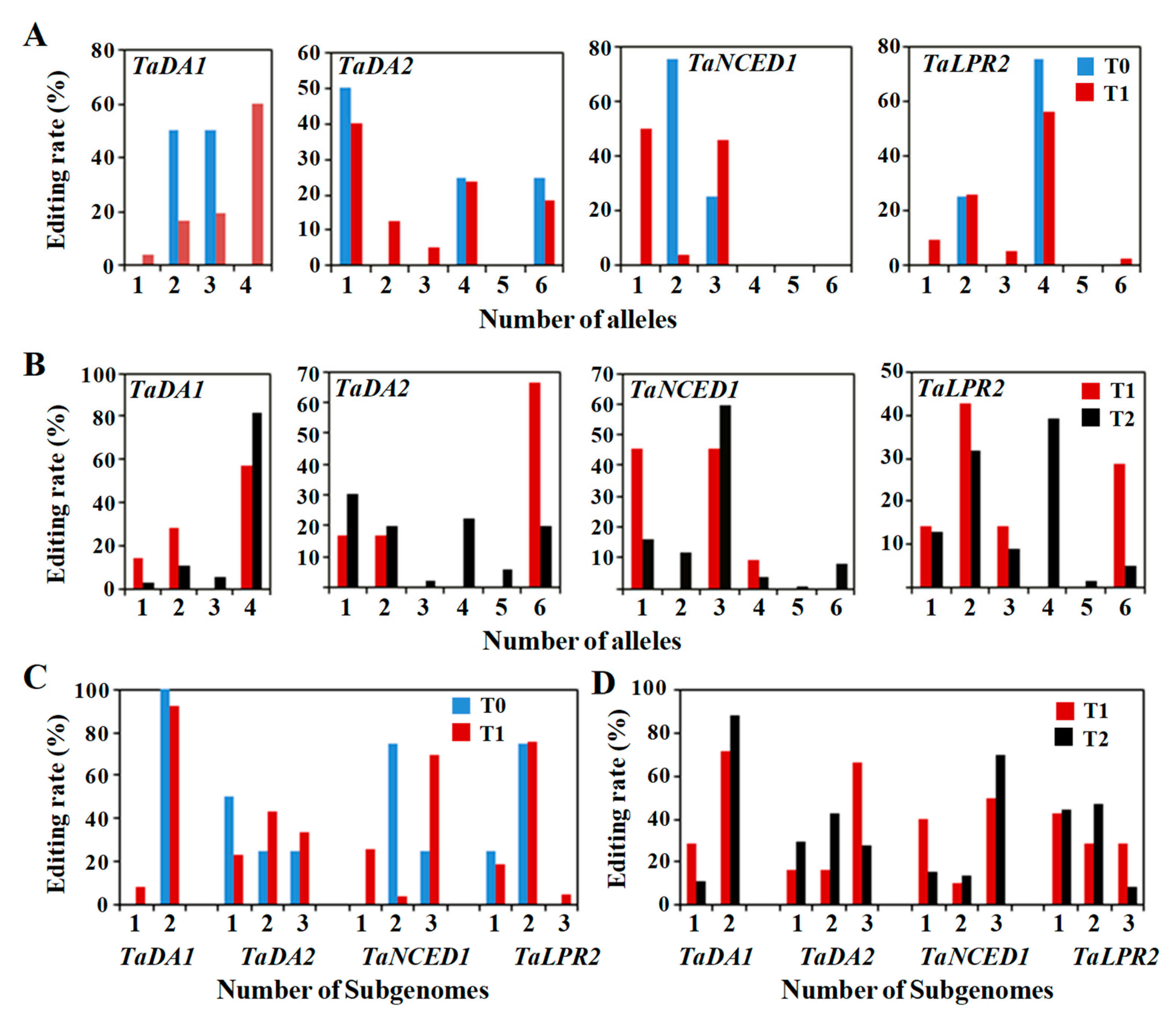
2.5. Editing Frequency of the Mutant Genotypes across the Generations
2.6. Off-Target Analysis of Targeting sgRNAs
3. Discussion
4. Material and Methods
4.1. sgRNA Design and Plasmid Construction
4.2. Plant Materials and Genetic Transformation
4.3. Detection of the Targeted Genome Editing Events
4.4. Off-Target Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DSB | Double-Stranded Breaks |
| NHEJ | Non-Homologous End Joining |
| HDR | Homology-Directed Repair |
| PAM | Protospacer Adjacent Motif |
| TALEN | Transcription Activator-like Effector Nuclease |
| ZFN | Zinc Finger Nucleases |
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.G.; Liu, Y. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.6.1–31.6.21. [Google Scholar]
- Zhu, C.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of Genome Editing Mutations in Cereal Crops. Trends Plant Sci. 2017, 22, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Bao, A.; Chen, H.; Chen, L.; Chen, S.; Hao, Q.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K.; et al. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 2017, 15, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Yuan, J.; Wang, R.; Liu, Y.; Birchler, J.A.; Han, F. Efficient Targeted Genome Modification in Maize Using CRISPR/Cas9 System. J. Genet. Genom. 2016, 43, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wang, N. Targeted Genome Editing of Sweet Orange Using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef] [PubMed]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, X.; Xie, X.; Liu, Y.-G. CRISPR/Cas9-Based Genome Editing in Plants. Prog. Mol. Biol. Transl. Sci. 2017, 149, 133–150. [Google Scholar]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- Ling, H.Q.; Zhao, S.; Liu, D.; Wang, J.; Sun, H.; Zhang, C.; Fan, H.; Li, D.; Dong, L.; Tao, Y.; et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 2013, 496, 87–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Leon, S.; Gil-Humanes, J.; Ozuna, C.V.; Gimenez, M.J.; Sousa, C.; Voytas, D.F.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Zhang, D.; Zhao, X.; Cao, X.; Dong, L.; Liu, J.; Chen, K.; Zhang, H.; Gao, C.; et al. Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 2018, 94, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Howells, R.M.; Craze, M.; Bowden, S.; Wallington, E.J. Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biol. 2018, 18, 215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, R.; Song, G.; Gao, J.; Li, W.; Han, X.; Chen, M.; Li, Y.; Li, G. Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biol. 2018, 18, 302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 2019, 17, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, M.; Thilges, K.; Cho, M.J.; Cigan, A.M. MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum L.) and combining mutations in all three homeologs causes male sterility. PLoS ONE 2017, 12, e0177632. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.S.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR /Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Song, N.; Sun, S.; Yang, W.; Zhao, H.; Song, W.; Lai, J. Efficiency and Inheritance of Targeted Mutagenesis in Maize Using CRISPR-Cas9. J. Genet. Genom. 2016, 43, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Liu, Y.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 2018, 8, 14422. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Simmonds, J.; Pan, Q.; Davidson, D.; He, F.; Battal, A.; Akhunova, A.; Trick, H.N.; Uauy, C.; Akhunov, E. Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 2018, 131, 2463–2475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- Morineau, C.; Bellec, Y.; Tellier, F.; Gissot, L.; Kelemen, Z.; Nogué, F.; Faure, J.D. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 2017, 15, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Wang, Z.P.; Xing, H.L.; Dong, L.; Zhang, H.Y.; Han, C.Y.; Wang, X.C.; Chen, Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 905. [Google Scholar] [CrossRef]
- Daer, R.M.; Cutts, J.P.; Brafman, D.A.; Haynes, K.A. The Impact of Chromatin Dynamics on Cas9-Mediated Genome Editing in Human Cells. ACS Synth. Biol. 2017, 6, 428–438. [Google Scholar] [CrossRef]
- Kallimasioti-Pazi, E.M.; Chathoth, K.T.; Taylor, G.C.; Meynert, A.; Ballinger, T.; Kelder, M.J.E.; Lalevée, S.; Sanli, I.; Feil, R.; Wood, A.J. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLOS Biol. 2018, 16, e2005595. [Google Scholar] [CrossRef] [PubMed]
- Uusi-Mäkelä, M.I.E.; Barker, H.R.; Bäuerlein, C.A.; Hakkinen, T.; Nykter, M.; Rämet, M. Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio). PLoS ONE 2018, 13, e0196238. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Number of Plants Examined | Mutation Line | Mutation Rate (%) | Mutation Types (bp) |
|---|---|---|---|---|
| TaPinb | 22 | 0 | 0 | - |
| TaDA1 | 24 | 13 | 54.2 | 1i, 1d, 2d, 4d, 5d, 19d |
| TaDA2 | 19 | 6 | 31.2 | 1i, 1d |
| TaNCED1 | 48 | 10 | 20.8 | 1i, 1d, 2d, 8d |
| TaLPR2 | 15 | 7 | 46.7 | 1i, 1d, 3d, 4d, 6d, 29d, 42d |
| Target gene | T1 Line | Number of Plants Examined | Number of Mutated Plants | Mutation Rates (%) | Mutation Types (bp) | T2 Line from T1 | Number of Plants Examined | Number of Mutated Plants | Mutation Rates (%) | Mutation Type (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| TaPinb | 1-22 | 132 | 10 | 7.6% | 5d, 7d, 12d, 29d, 34d | 47 | 21 | 1 | 4.76 | 12d |
| 30 | 21 | 15 | 71.4 | 12d | ||||||
| 34 | 21 | 2 | 9.5 | 12d | ||||||
| 44 | 21 | 2 | 9.5 | 12d | ||||||
| 48 | 12 | 1 | 8.3 | 12d | ||||||
| TaDA1 | 6 | 13 | 12 | 92.3 | 1d, 3d, 1i | 6-3 | 17 | 17 | 100 | 1i, 1d, 3d, 6d |
| 14 | 21 | 20 | 95.2 | 4d, 5d,7d, 9d, 18d, 1i | 14-1 | 19 | 19 | 100 | 1i, 4d, 5d, 11d, | |
| 14-15 | 19 | 19 | 100 | 5d, 9d, 18d | ||||||
| 14-6 | 19 | 18 | 94.7 | 1i, 4d, 5d, 9d, 18d | ||||||
| 15 | 25 | 25 | 100 | 1d, 2d, 3d, 4d, 1i | 15-7 | 15 | 15 | 100 | 1i, 1d, 2d, 3d, 4d | |
| 15-8 | 15 | 15 | 100 | 1i, 2d | ||||||
| 17 | 26 | 26 | 100 | 1d, 2d, 4d, 12d, 19d, 57d, 1i | 17-4 | 16 | 16 | 100 | 1i, 1d, 2d | |
| TaDA2 | 3 | 19 | 15 | 78.9 | 53d, 1i | 3-12 | 17 | 13 | 76.5 | 1i |
| 3-13 | 13 | 12 | 92.3 | 1i | ||||||
| 5 | 13 | 8 | 61.5 | 1d, 1i | 5-13 | 22 | 16 | 72.7 | 1i | |
| 58 | 20 | 18 | 90 | 1d, 7d, 1i | 58-6 | 21 | 21 | 100 | 1i, 1d | |
| 58-10 | 16 | 16 | 100 | 1i, 1d | ||||||
| 58-18 | 12 | 12 | 100 | 1i, 1d | ||||||
| 60 | 22 | 14 | 63.6 | 1d, 24d, 54d, 1i | 60-3 | 14 | 1 | 7.1 | 1d | |
| TaNCED1 | 53 | 11 | 7 | 63.6 | 1d, 8d | 53-1 | 18 | 16 | 88.9 | 1d, 8d |
| 53-6 | 20 | 20 | 100 | 1d, 8d | ||||||
| 53-10 | 15 | 15 | 100 | 8d | ||||||
| 84 | 20 | 14 | 70 | 1d | 84-6 | 20 | 10 | 50.0 | 1d | |
| 84-10 | 14 | 14 | 100 | 1d | ||||||
| 98 | 17 | 16 | 94.1 | 1i, 12d | 98-3 | 8 | 8 | 100 | 1i | |
| 98-9 | 16 | 16 | 100 | 1i, 1d | ||||||
| 98-12 | 18 | 14 | 77.8 | 1i | ||||||
| 98-13 | 11 | 11 | 100 | 1i, 12d | ||||||
| 107 | 16 | 11 | 68.8 | 2d | 107-2 | 11 | 3 | 27.3 | 2d | |
| TaLPR2 | 1 | 22 | 22 | 100 | 1d, 4d, 6d, 2i | 1-13 | 20 | 20 | 100 | 1d, 4d, 6d |
| 54 | 23 | 23 | 100 | 1d, 12d | 54-23 | 20 | 20 | 100 | 1d | |
| 57 | 18 | 14 | 77.8 | 29d, 1i | 57-1 | 18 | 13 | 72.2 | 1i | |
| 59 | 21 | 14 | 66.7 | 1d, 2d, 6d, 12d, 13d, 15d, 18d, 22d, 30d, 50d, 53d, 1i | 59-4 | 16 | 16 | 100 | 1d, 1i, 2d, 6d, 9d, 12d, 17d | |
| 59-11 | 20 | 20 | 100 | 1d, 3d, 5d, 22d, 93d2i | ||||||
| 59-14 | 15 | 15 | 100 | 1d, 2d, 4d, 6d | ||||||
| 59-16 | 18 | 18 | 100 | 1d, 4d, 6d, 7d, 12d, 18d, 30d |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Zhang, R.; Gao, J.; Gu, T.; Song, G.; Li, W.; Li, D.; Li, Y.; Li, G. Highly Efficient and Heritable Targeted Mutagenesis in Wheat via the Agrobacterium tumefaciens-Mediated CRISPR/Cas9 System. Int. J. Mol. Sci. 2019, 20, 4257. https://doi.org/10.3390/ijms20174257
Zhang S, Zhang R, Gao J, Gu T, Song G, Li W, Li D, Li Y, Li G. Highly Efficient and Heritable Targeted Mutagenesis in Wheat via the Agrobacterium tumefaciens-Mediated CRISPR/Cas9 System. International Journal of Molecular Sciences. 2019; 20(17):4257. https://doi.org/10.3390/ijms20174257
Chicago/Turabian StyleZhang, Shujuan, Rongzhi Zhang, Jie Gao, Tiantian Gu, Guoqi Song, Wei Li, Dandan Li, Yulian Li, and Genying Li. 2019. "Highly Efficient and Heritable Targeted Mutagenesis in Wheat via the Agrobacterium tumefaciens-Mediated CRISPR/Cas9 System" International Journal of Molecular Sciences 20, no. 17: 4257. https://doi.org/10.3390/ijms20174257
APA StyleZhang, S., Zhang, R., Gao, J., Gu, T., Song, G., Li, W., Li, D., Li, Y., & Li, G. (2019). Highly Efficient and Heritable Targeted Mutagenesis in Wheat via the Agrobacterium tumefaciens-Mediated CRISPR/Cas9 System. International Journal of Molecular Sciences, 20(17), 4257. https://doi.org/10.3390/ijms20174257