Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice
Abstract
:1. Introduction
2. Results
2.1. Nobiletin Improves Serum Cholesterol Profiles in Aged Mice under High-Fat Feeding
2.2. Nobiletin Attenuates Bile Acids Leakage in the Serum and Safeguards Healthy Liver
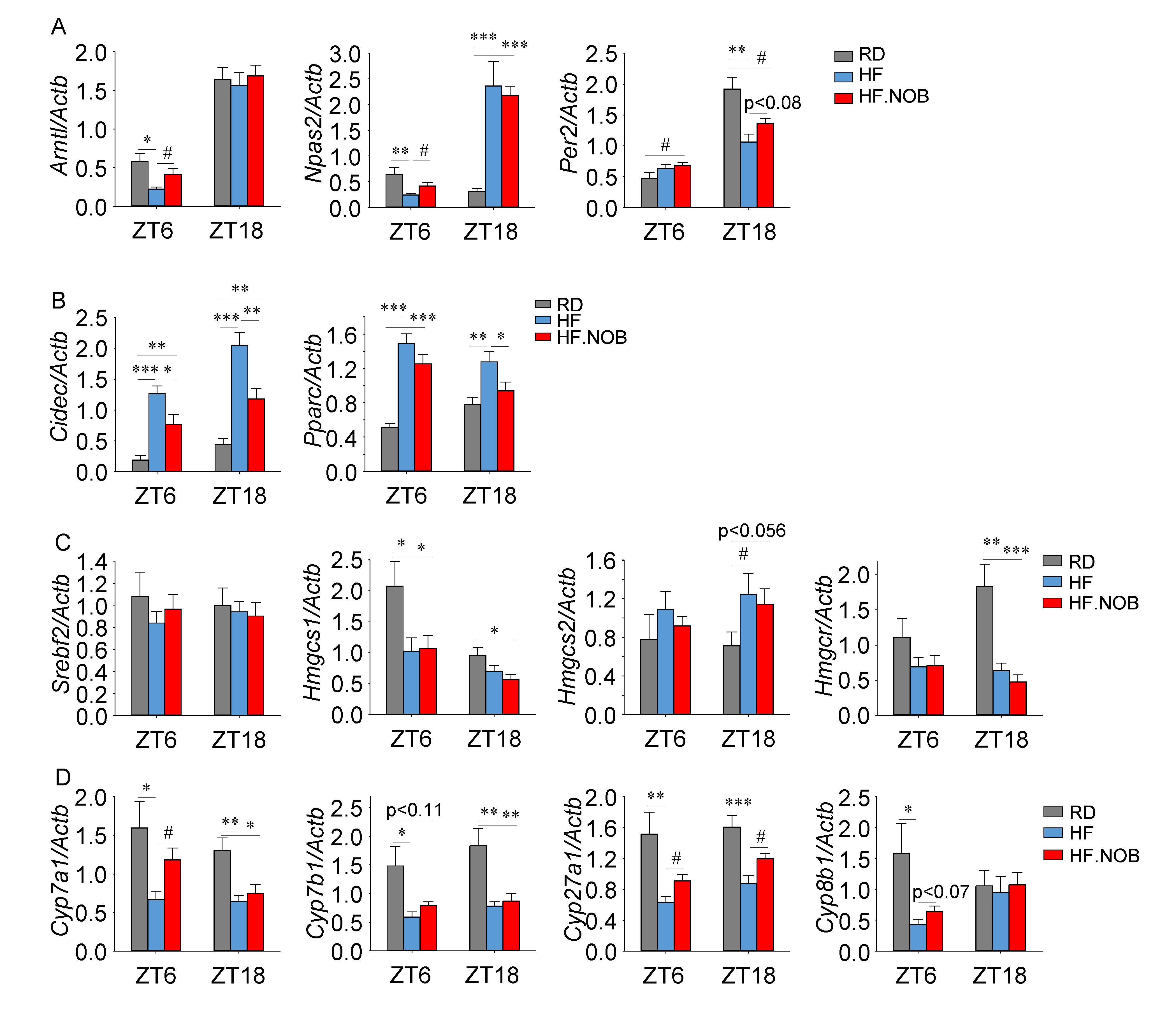
2.3. Nobiletin Reprograms Circadian and Lipid Homeostasis Genes in the Liver
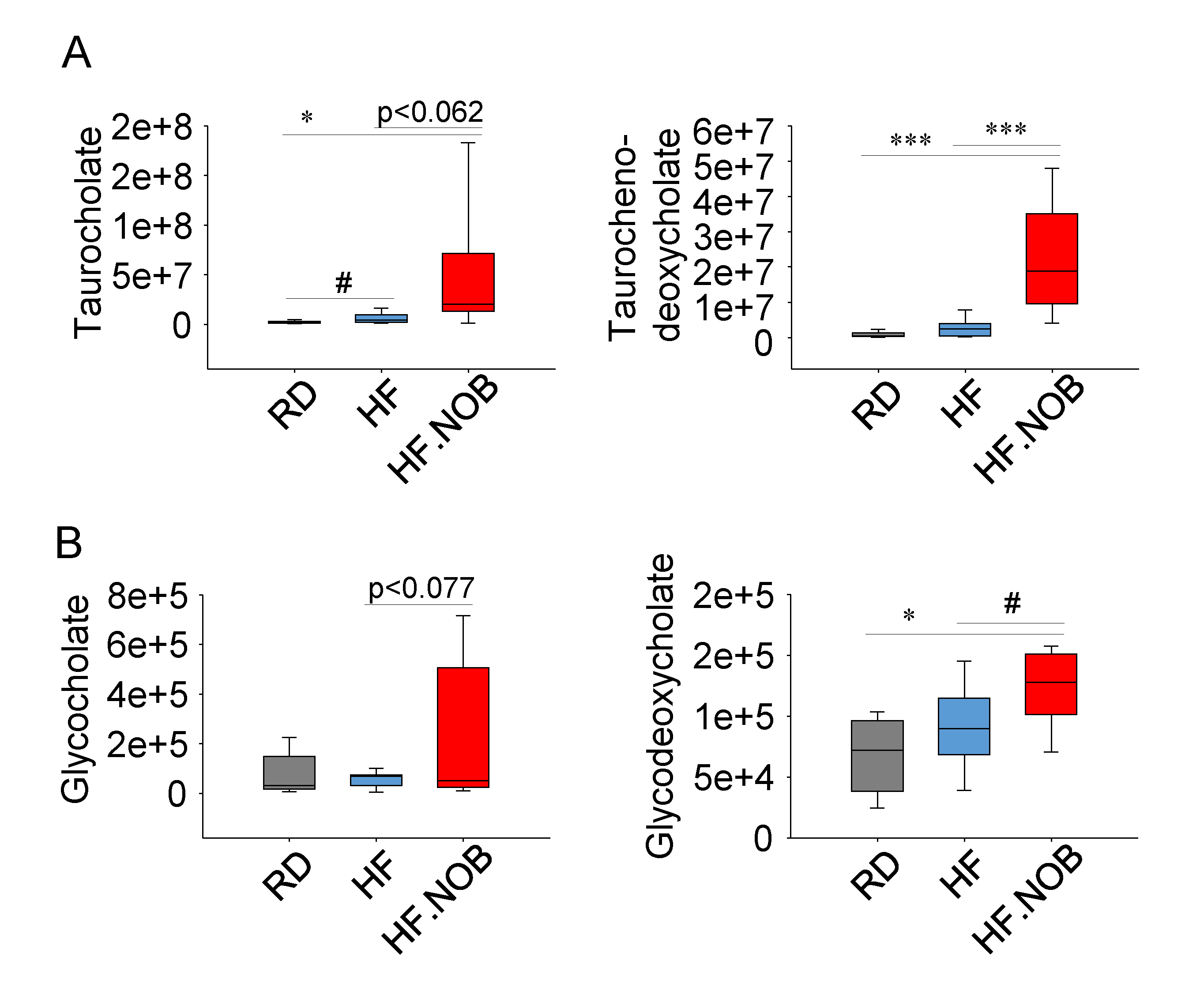
2.4. Nobiletin Upregulates Fecal Bile Acids Levels
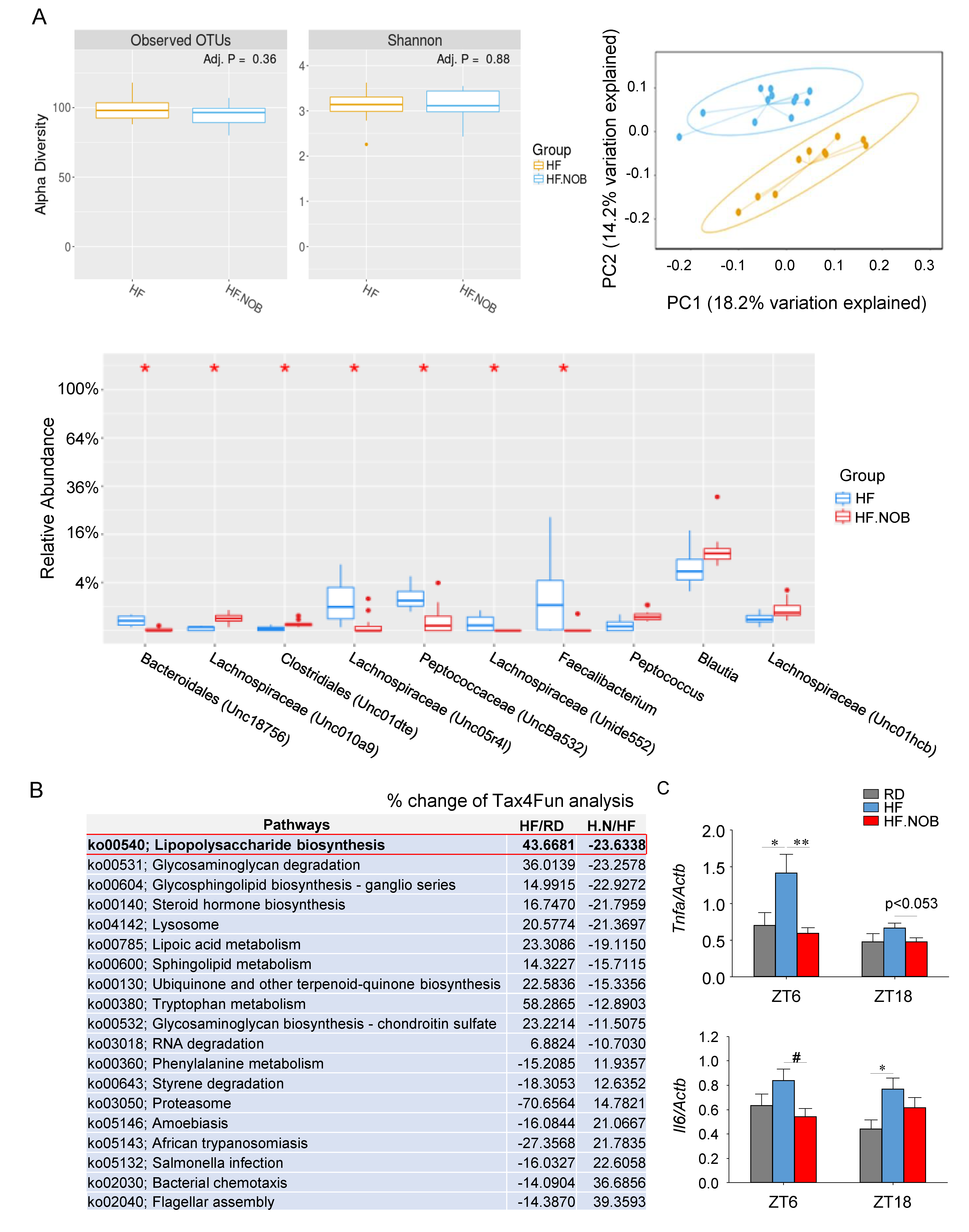
2.5. Nobiletin Remodels Gut Microbiota
3. Discussion
4. Materials and Methods
4.1. Animal Studies
4.2. Serum Content Assays
4.3. Real-Time qPCR
4.4. Metabolomic Analysis
4.4.1. Sample Preparation
4.4.2. UHPLC-MS Analysis
4.5. Microbiome 16S Sequencing
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goldstein, J.L.; Brown, M.S. A century of cholesterol and coronaries: From plaques to genes to statins. Cell 2015, 161, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Chiang JY, L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu Rev. Nutr 2019. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Reviews. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Wahlstrom, A.; Sayin, S.I.; Marschall, H.U.; Backhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Albrecht, U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.C.; Welsh, D.K.; Ko, C.H.; Tran, H.G.; Zhang, E.E.; Priest, A.A.; Buhr, E.D.; Singer, O.; Meeker, K.; Verma, I.M.; et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 2007, 129, 605–616. [Google Scholar] [CrossRef]
- Challet, E. The circadian regulation of food intake. Nat. Reviews. Endocrinol. 2019, 15, 393–405. [Google Scholar] [CrossRef]
- Bass, J.; Lazar, M.A. Circadian time signatures of fitness and disease. Science 2016, 354, 994–999. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [Green Version]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 16, 359. [Google Scholar] [CrossRef] [PubMed]
- Paschos, G.K.; FitzGerald, G.A. Circadian Clocks and Metabolism: Implications for Microbiome and Aging. Trends Genet. 2017, 33, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalova, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E.; et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 2016, 167, 1495–1510. [Google Scholar] [CrossRef] [PubMed]
- Weger, B.D.; Gobet, C.; Yeung, J.; Martin, E.; Jimenez, S.; Betrisey, B.; Foata, F.; Berger, B.; Balvay, A.; Foussier, A.; et al. The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab. 2019, 29, 362–382. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Solanas, G.; Peixoto, F.O.; Bee, L.; Symeonidi, A.; Schmidt, M.S.; Brenner, C.; Masri, S.; Benitah, S.A.; Sassone-Corsi, P. Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 2017, 170, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Straume, M.; Tei, H.; Sakaki, Y.; Menaker, M.; Block, G.D. Effects of aging on central and peripheral mammalian clocks. Proc. Natl. Acad. Sci. USA 2002, 99, 10801–10806. [Google Scholar] [CrossRef] [Green Version]
- Tahara, Y.; Takatsu, Y.; Shiraishi, T.; Kikuchi, Y.; Yamazaki, M.; Motohashi, H.; Muto, A.; Sasaki, H.; Haraguchi, A.; Kuriki, D.; et al. Age-related circadian disorganization caused by sympathetic dysfunction in peripheral clock regulation. Npj Aging Mech. Dis. 2017, 3, 16030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca Costa, S.S.; Ripperger, J.A. Impact of the circadian clock on the aging process. Front. Neurol. 2015, 6, 43. [Google Scholar] [CrossRef]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [Green Version]
- Davidson, A.J.; Sellix, M.T.; Daniel, J.; Yamazaki, S.; Menaker, M.; Block, G.D. Chronic jet-lag increases mortality in aged mice. Curr. Biol. 2006, 16, R914–R916. [Google Scholar] [CrossRef] [Green Version]
- Adamovich, Y.; Rousso-Noori, L.; Zwighaft, Z.; Neufeld-Cohen, A.; Golik, M.; Kraut-Cohen, J.; Wang, M.; Han, X.; Asher, G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014, 19, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Zmrzljak, U.P.; Rozman, D. Circadian regulation of the hepatic endobiotic and xenobitoic detoxification pathways: The time matters. Chem. Res. Toxicol. 2012, 25, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.K.; Guo, G.L.; Klaassen, C.D. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE 2011, 6, e16683. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Voicu, H.; Finegold, M.J.; Coarfa, C.; Sreekumar, A.; Putluri, N.; Katchy, C.A.; Lee, C.; Moore, D.D.; Fu, L. Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 2016, 30, 909–924. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhang, R.; Jain, R.; Shi, H.; Zhang, L.; Zhou, G.; Sangwung, P.; Tugal, D.; Atkins, G.B.; Prosdocimo, D.A.; et al. Circadian control of bile acid synthesis by a KLF15-Fgf15 axis. Nat. Commun. 2015, 6, 7231. [Google Scholar] [CrossRef] [Green Version]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Shostak, A.; Meyer-Kovac, J.; Oster, H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes 2013, 62, 2195–2203. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef]
- Ma, K.; Xiao, R.; Tseng, H.T.; Shan, L.; Fu, L.; Moore, D.D. Circadian dysregulation disrupts bile acid homeostasis. PLoS ONE 2009, 4, e6843. [Google Scholar] [CrossRef]
- Ferrell, J.M.; Chiang, J.Y. Short-term circadian disruption impairs bile acid and lipid homeostasis in mice. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 664–677. [Google Scholar] [CrossRef]
- Yamajuku, D.; Okubo, S.; Haruma, T.; Inagaki, T.; Okuda, Y.; Kojima, T.; Noutomi, K.; Hashimoto, S.; Oda, H. Regular feeding plays an important role in cholesterol homeostasis through the liver circadian clock. Circ. Res. 2009, 105, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Bradfield, C.A.; Hussain, M.M. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat. Commun. 2016, 7, 13011. [Google Scholar] [CrossRef] [PubMed]
- Le Martelot, G.; Claudel, T.; Gatfield, D.; Schaad, O.; Kornmann, B.; Lo Sasso, G.; Moschetta, A.; Schibler, U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. Plos Biol. 2009, 7, e1000181. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.; Fitzsimmons, R.L.; Raichur, S.; Wang, S.C.; Lechtken, A.; Muscat, G.E. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: Staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008, 283, 18411–18421. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Kang, H.S.; Lih, F.B.; Jiang, H.; Blaner, W.S.; Jetten, A.M. Retinoid acid-related orphan receptor gamma, RORgamma, participates in diurnal transcriptional regulation of lipid metabolic genes. Nucleic Acids Res. 2014, 42, 10448–10459. [Google Scholar] [CrossRef]
- Kang, H.S.; Angers, M.; Beak, J.Y.; Wu, X.; Gimble, J.M.; Wada, T.; Xie, W.; Collins, J.B.; Grissom, S.F.; Jetten, A.M. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol. Genom. 2007, 31, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Kang, H.S.; Angers, M.; Gong, H.; Bhatia, S.; Khadem, S.; Ren, S.; Ellis, E.; Strom, S.C.; Jetten, A.M.; et al. Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor alpha (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3). Mol. Pharm. 2008, 73, 891–899. [Google Scholar] [CrossRef]
- Kim, D.H.; Rhee, J.C.; Yeo, S.; Shen, R.; Lee, S.K.; Lee, J.W.; Lee, S. Crucial roles of mixed-lineage leukemia 3 and 4 as epigenetic switches of the hepatic circadian clock controlling bile acid homeostasis in mice. Hepatology 2015, 61, 1012–1023. [Google Scholar] [CrossRef]
- Pathak, P.; Li, T.; Chiang, J.Y. Retinoic acid-related orphan receptor alpha regulates diurnal rhythm and fasting induction of sterol 12alpha-hydroxylase in bile acid synthesis. J. Biol. Chem. 2013, 288, 37154–37165. [Google Scholar] [CrossRef]
- Soroosh, P.; Wu, J.; Xue, X.; Song, J.; Sutton, S.W.; Sablad, M.; Yu, J.; Nelen, M.I.; Liu, X.; Castro, G.; et al. Oxysterols are agonist ligands of RORgammat and drive Th17 cell differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, 12163–12168. [Google Scholar] [CrossRef]
- Kojetin, D.J.; Burris, T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discov. 2014, 13, 197–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santori, F.R.; Huang, P.; van de Pavert, S.A.; Douglass, E.F., Jr.; Leaver, D.J.; Haubrich, B.A.; Keber, R.; Lorbek, G.; Konijn, T.; Rosales, B.N.; et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab. 2015, 21, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kumar, N.; Solt, L.A.; Richardson, T.I.; Helvering, L.M.; Crumbley, C.; Garcia-Ordonez, R.D.; Stayrook, K.R.; Zhang, X.; Novick, S.; et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J. Biol. Chem. 2010, 285, 5013–5025. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Nohara, K.; Park, N.; Park, Y.S.; Guillory, B.; Zhao, Z.; Garcia, J.M.; Koike, N.; Lee, C.C.; Takahashi, J.S.; et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab. 2016, 23, 610–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nohara, K.; Shin, Y.; Park, N.; Jeong, K.; He, B.; Koike, N.; Yoo, S.H.; Chen, Z. Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid. Nutr. Metab. (Lond.) 2015, 12, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nohara, K.; Mallampalli, V.; Nemkov, T.; Wirianto, M.; Yang, J.; Ye, Y.; Sun, Y.; Han, L.; Esser, K.A.; Mileykovskaya, E.; et al. Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat. Commun. 2019, 10, 3923. [Google Scholar] [CrossRef] [PubMed]
- Manninen, V.; Tenkanen, L.; Koskinen, P.; Huttunen, J.K.; Manttari, M.; Heinonen, O.P.; Frick, M.H. Joint effects of serum triglyceride and LDL cholesterol and HDL cholesterol concentrations on coronary heart disease risk in the Helsinki Heart Study. Implications for treatment. Circulation 1992, 85, 37–45. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef]
- McGill, M.R.; Li, F.; Sharpe, M.R.; Williams, C.D.; Curry, S.C.; Ma, X.; Jaeschke, H. Circulating acylcarnitines as biomarkers of mitochondrial dysfunction after acetaminophen overdose in mice and humans. Arch. Toxicol. 2014, 88, 391–401. [Google Scholar] [CrossRef]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 1998, 101, 2331–2339. [Google Scholar] [CrossRef]
- Huang, H.; Li, L.; Shi, W.; Liu, H.; Yang, J.; Yuan, X.; Wu, L. The Multifunctional Effects of Nobiletin and Its Metabolites In Vivo and In Vitro. Evid Based Complement. Altern. Med. 2016, 2016, 2918796. [Google Scholar] [CrossRef] [PubMed]
- Mulvihill, E.E.; Burke, A.C.; Huff, M.W. Citrus Flavonoids as Regulators of Lipoprotein Metabolism and Atherosclerosis. Annu Rev. Nutr. 2016, 36, 275–299. [Google Scholar] [CrossRef] [PubMed]
- Gloston, G.F.; Yoo, S.H.; Chen, Z.J. Clock-Enhancing Small Molecules and Potential Applications in Chronic Diseases and Aging. Front. Neurol. 2017, 8, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulvihill, E.E.; Assini, J.M.; Lee, J.K.; Allister, E.M.; Sutherland, B.G.; Koppes, J.B.; Sawyez, C.G.; Edwards, J.Y.; Telford, D.E.; Charbonneau, A.; et al. Nobiletin attenuates VLDL overproduction, dyslipidemia, and atherosclerosis in mice with diet-induced insulin resistance. Diabetes 2011, 60, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, A.; Misawa, K.; Ikeda, Y.; Haraguchi, A.; Kamagata, M.; Tahara, Y.; Shibata, S. Potent Effects of Flavonoid Nobiletin on Amplitude, Period, and Phase of the Circadian Clock Rhythm in PER2::LUCIFERASE Mouse Embryonic Fibroblasts. PLoS ONE 2017, 12, e0170904. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Guo, R.; Tian, H.; Li, L.; Liu, H.; Mi, Y.; Liu, X. Nobiletin protects against insulin resistance and disorders of lipid metabolism by reprogramming of circadian clock in hepatocytes. Biochim. Et Biophys. Acta. Mol. Cell Biol. Lipids 2018, 1863, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Gile, J.; Scott, B.; Eckle, T. The Period 2 Enhancer Nobiletin as Novel Therapy in Murine Models of Circadian Disruption Resembling Delirium. Crit. Care Med. 2018, 46, e600–e608. [Google Scholar] [CrossRef] [PubMed]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Galluzzi, L.; Freije, J.M.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef] [Green Version]
- Manoogian, E.N.C.; Panda, S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev. 2017, 39, 59–67. [Google Scholar] [CrossRef]
- Patel, S.A.; Chaudhari, A.; Gupta, R.; Velingkaar, N.; Kondratov, R.V. Circadian clocks govern calorie restriction-mediated life span extension through BMAL1- and IGF-1-dependent mechanisms. Faseb J. 2016, 30, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Head, T.; Daunert, S.; Goldschmidt-Clermont, P.J. The Aging Risk and Atherosclerosis: A Fresh Look at Arterial Homeostasis. Front. Genet. 2017, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Einarsson, K.; Nilsell, K.; Leijd, B.; Angelin, B. Influence of age on secretion of cholesterol and synthesis of bile acids by the liver. N. Engl. J. Med. 1985, 313, 277–282. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Chen, Z. Molecular Targets for Small-Molecule Modulators of Circadian Clocks. Curr. Drug Metab. 2016, 17, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallach, T.; Kramer, A. Chemical chronobiology: Toward drugs manipulating time. Febs. Lett. 2015, 589, 1530–1538. [Google Scholar] [CrossRef] [Green Version]
- Isojima, Y.; Nakajima, M.; Ukai, H.; Fujishima, H.; Yamada, R.G.; Masumoto, K.H.; Kiuchi, R.; Ishida, M.; Ukai-Tadenuma, M.; Minami, Y.; et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 15744–15749. [Google Scholar] [CrossRef] [PubMed]
- Hirota, T.; Kay, S.A. High-throughput screening and chemical biology: New approaches for understanding circadian clock mechanisms. Chem. Biol. 2009, 16, 921–927. [Google Scholar] [CrossRef]
- Cha, H.K.; Chung, S.; Lim, H.Y.; Jung, J.W.; Son, G.H. Small Molecule Modulators of the Circadian Molecular Clock With Implications for Neuropsychiatric Diseases. Front. Mol. Neurosci. 2018, 11, 496. [Google Scholar] [CrossRef]
- Solt, L.A.; Wang, Y.; Banerjee, S.; Hughes, T.; Kojetin, D.J.; Lundasen, T.; Shin, Y.; Liu, J.; Cameron, M.D.; Noel, R.; et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012, 485, 62–68. [Google Scholar] [CrossRef]
- Chang, M.R.; He, Y.; Khan, T.M.; Kuruvilla, D.S.; Garcia-Ordonez, R.; Corzo, C.A.; Unger, T.J.; White, D.W.; Khan, S.; Lin, L.; et al. Antiobesity Effect of a Small Molecule Repressor of RORgamma. Mol. Pharm. 2015, 88, 48–56. [Google Scholar] [CrossRef]
- Oshima, T.; Niwa, Y.; Kuwata, K.; Srivastava, A.; Hyoda, T.; Tsuchiya, Y.; Kumagai, M.; Tsuyuguchi, M.; Tamaru, T.; Sugiyama, A.; et al. Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 2019, 5, eaau9060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W.; et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Lee, E.J.; Yun, S.; Choe, H.K.; Park, S.B.; Son, H.J.; Kim, K.S.; Dluzen, D.E.; Lee, I.; Hwang, O.; et al. Impact of circadian nuclear receptor REV-ERBalpha on midbrain dopamine production and mood regulation. Cell 2014, 157, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Sulli, G.; Rommel, A.; Wang, X.; Kolar, M.J.; Puca, F.; Saghatelian, A.; Plikus, M.V.; Verma, I.M.; Panda, S. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 2018, 553, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Panda, S. Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell Metab. 2016, 23, 1048–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, S.; Basse, A.L.; Schonke, M.; Chen, S.; Samad, M.; Altintas, A.; Laker, R.C.; Dalbram, E.; Barres, R.; Baldi, P.; et al. Time of Exercise Specifies the Impact on Muscle Metabolic Pathways and Systemic Energy Homeostasis. Cell Metab. 2019, 30, 92–110. [Google Scholar] [CrossRef]
- Chen, Z.; Yoo, S.H.; Takahashi, J.S. Development and Therapeutic Potential of Small-Molecule Modulators of Circadian Systems. Annu. Rev. Pharm. Toxicol. 2018, 58, 231–252. [Google Scholar] [CrossRef]
- Schroeder, A.M.; Colwell, C.S. How to fix a broken clock. Trends Pharm. Sci. 2013, 34, 605–619. [Google Scholar] [CrossRef] [Green Version]
- Montaigne, D.; Marechal, X.; Modine, T.; Coisne, A.; Mouton, S.; Fayad, G.; Ninni, S.; Klein, C.; Ortmans, S.; Seunes, C.; et al. Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbalpha antagonism: A single-centre propensity-matched cohort study and a randomised study. Lancet 2018, 391, 59–69. [Google Scholar] [CrossRef]
- Ruben, M.D.; Smith, D.F.; FitzGerald, G.A.; Hogenesch, J.B. Dosing time matters. Science 2019, 365, 547–549. [Google Scholar] [CrossRef]
- Cederroth, C.R.; Albrecht, U.; Bass, J.; Brown, S.A.; Dyhrfjeld-Johnsen, J.; Gachon, F.; Green, C.B.; Hastings, M.H.; Helfrich-Forster, C.; Hogenesch, J.B.; et al. Medicine in the Fourth Dimension. Cell Metab. 2019, 30, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. What’s next for chronobiology and drug discovery. Expert Opin. Drug Discov. 2017, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Sutton, E.F.; Beyl, R.; Early, K.S.; Cefalu, W.T.; Ravussin, E.; Peterson, C.M. Early Time-Restricted Feeding Improves Insulin Sensitivity, Blood Pressure, and Oxidative Stress Even without Weight Loss in Men with Prediabetes. Cell Metab 2018, 27, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.; Sharma, P.; Guthrie, N. Bioavailability of Citrus Polymethoxylated Flavones and Their Biological Role in Metabolic Syndrome and Hyperlipidemia. InTech 2012, 267–284. [Google Scholar] [CrossRef]
- Nemkov, T.; Hansen, K.C.; D’Alessandro, A. A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun. Mass Spectrom. 2017, 31, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Clasquin, M.F.; Melamud, E.; Rabinowitz, J.D. LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Curr. Protoc. Bioinform. 2012, 37, 14.11.1–14.11.23. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Nohara, K.; Ajami, N.J.; Michalek, R.D.; Tian, X.; Wong, M.; Losee-Olson, S.H.; Petrosino, J.F.; Yoo, S.H.; Shimomura, K.; et al. Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis. Sci. Rep. 2015, 5, 10604. [Google Scholar] [CrossRef]
- Asshauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nohara, K.; Nemkov, T.; D’Alessandro, A.; Yoo, S.-H.; Chen, Z. Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice. Int. J. Mol. Sci. 2019, 20, 4281. https://doi.org/10.3390/ijms20174281
Nohara K, Nemkov T, D’Alessandro A, Yoo S-H, Chen Z. Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice. International Journal of Molecular Sciences. 2019; 20(17):4281. https://doi.org/10.3390/ijms20174281
Chicago/Turabian StyleNohara, Kazunari, Travis Nemkov, Angelo D’Alessandro, Seung-Hee Yoo, and Zheng Chen. 2019. "Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice" International Journal of Molecular Sciences 20, no. 17: 4281. https://doi.org/10.3390/ijms20174281
APA StyleNohara, K., Nemkov, T., D’Alessandro, A., Yoo, S.-H., & Chen, Z. (2019). Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice. International Journal of Molecular Sciences, 20(17), 4281. https://doi.org/10.3390/ijms20174281




