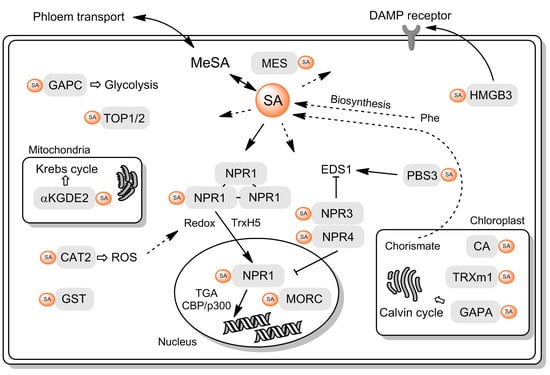
Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling
Abstract
:1. Introduction
2. SA Binding Proteins
2.1. SABP1—Catalase
2.2. SABP2—MeSA Esterase
2.3. SABP3—β Carbonic Anhydrase
2.4. NPR1/2/3/4—Signalling Proteins
2.5. Glutathione S-transferase
2.6. Thioredoxins
2.7. GH3—Acyl Acid Amido Synthetase
2.8. GAPDH
2.9. Alpha-ketoglutarate Dehydrogenase—Krebs Cycle Enzyme
2.10. Thimet Oligopeptidases + TPPII Exopeptidase—Proteolysis
2.11. MORC Proteins—Epigenetic Regulation
2.12. HMGB3—DAMP Protein
3. Response of SABPs to Treatments Linked to SA/Biotic Stress
4. Molecular Mechanisms of SA-Protein Interactions
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dong, C.-J.; Li, L.; Shang, Q.-M.; Liu, X.-Y.; Zhang, Z.-G. Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta 2014, 240, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Min, K.; Arora, R. Exogenous salicylic acid improves freezing tolerance of spinach (Spinacia oleracea L.) leaves. Cryobiology 2018, 81, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Cao, W.; Jiang, D. Hydrogen Peroxide and Abscisic Acid Mediate Salicylic Acid-Induced Freezing Tolerance in Wheat. Front. Plant. Sci. 2018, 9, 1137. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Zhang, C.; Chen, T.; Zhang, X.; Tao, L.; Fu, G. Salicylic acid reverses pollen abortion of rice caused by heat stress. BMC Plant Biol. 2018, 18, 245. [Google Scholar] [CrossRef] [PubMed]
- Moravcová, Š.; Tůma, J.; Dučaiová, Z.K.; Waligórski, P.; Kula, M.; Saja, D.; Słomka, A.; Bąba, W.; Libik-Konieczny, M. Influence of salicylic acid pretreatment on seeds germination and some defence mechanisms of Zea mays plants under copper stress. Plant Physiol. Biochem. 2018, 122, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Babourina, O.; Rengel, Z.; Jayakannan, M.; Bose, J.; Shabala, S.; Poschenrieder, C.; Massart, A. The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 1865–1875. [Google Scholar] [CrossRef]
- La, V.H.; Lee, B.-R.; Islam, M.T.; Park, S.-H.; Jung, H.-I.; Bae, D.-W.; Kim, T.-H. Characterization of salicylic acid-mediated modulation of the drought stress responses: Reactive oxygen species, proline, and redox state in Brassica napus. Environ. Exp. Bot. 2019, 157, 1–10. [Google Scholar] [CrossRef]
- Yang, L.; Li, B.; Zheng, X.-Y.; Li, J.; Yang, M.; Dong, X.; He, G.; An, C.; Deng, X.W. Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. Commun. 2015, 6, 7309. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Döring, A.-C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef]
- Prodhan, M.Y.; Munemasa, S.; Nahar, M.N.-E.-N.; Nakamura, Y.; Murata, Y. Guard Cell Salicylic Acid Signaling Is Integrated into Abscisic Acid Signaling via the Ca2+/CPK-Dependent Pathway. Plant Physiol. 2018, 178, 441–450. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.-G.; Park, C.-M. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010, 188, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Šašek, V.; Janda, M.; Delage, E.; Puyaubert, J.; Guivarc’h, A.; López Maseda, E.; Dobrev, P.I.; Caius, J.; Bóka, K.; Valentová, O.; et al. Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytol. 2014, 203, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Janda, M.; Ruelland, E. Magical mystery tour: Salicylic acid signalling. Environ. Exp. Bot. 2015, 114, 117–128. [Google Scholar] [CrossRef]
- Nawrath, C.; Métraux, J.-P. Salicylic Acid Induction-Deficient Mutants of Arabidopsis Express PR-2 and PR-5 and Accumulate High Levels of Camalexin after Pathogen Inoculation. Plant Cell 1999, 11, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562. [Google Scholar] [CrossRef] [PubMed]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Doermann, P.; Métraux, J.-P. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef]
- Hao, Q.; Wang, W.; Han, X.; Wu, J.; Lyu, B.; Chen, F.; Caplan, A.; Li, C.; Wu, J.; Wang, W.; et al. Isochorismate-based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 2018, 19, 1995–2010. [Google Scholar] [CrossRef]
- Seguel, A.; Jelenska, J.; Herrera-Vásquez, A.; Marr, S.K.; Joyce, M.B.; Gagesch, K.R.; Shakoor, N.; Jiang, S.-C.; Fonseca, A.; Wildermuth, M.C.; et al. PROHIBITIN3 Forms Complexes with ISOCHORISMATE SYNTHASE1 to Regulate Stress-Induced Salicylic Acid Biosynthesis in Arabidopsis. Plant Physiol. 2018, 176, 2515–2531. [Google Scholar] [CrossRef]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. bioRxiv 2019, 601948. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-X.; Zhang, W.; Dong, H.-X.; Liu, Z.-Y.; Ma, J.; Zhang, X.-Y. Salicylic acid in Populus tomentosa is a remote signalling molecule induced by Botryosphaeria dothidea infection. Sci. Rep. 2018, 8, 14059. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Vivancos, P.; Bernal-Vicente, A.; Cantabella, D.; Petri, C.; Hernández, J.A. Metabolomics and Biochemical Approaches Link Salicylic Acid Biosynthesis to Cyanogenesis in Peach Plants. Plant. Cell Physiol. 2017, 58, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.M.G.; Iancu, C.V.; Neet, K.E.; Dean, J.V.; Choe, J.-Y. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci. Rep. 2017, 7, 46629. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.V.; Mohammed, L.A.; Fitzpatrick, T. The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 2005, 221, 287–296. [Google Scholar] [CrossRef]
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.-J.; Gan, S.-S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef]
- Huang, X.-X.; Zhu, G.-Q.; Liu, Q.; Chen, L.; Li, Y.-J.; Hou, B.-K. Modulation of Plant Salicylic Acid-Associated Immune Responses via Glycosylation of Dihydroxybenzoic Acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar] [CrossRef]
- Baek, D.; Pathange, P.; Chung, J.-S.; Jiang, J.; Gao, L.; Oikawa, A.; Hirai, M.Y.; Saito, K.; Pare, P.W.; Shi, H. A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant Cell Environ. 2010, 33, 1383–1392. [Google Scholar] [CrossRef]
- Westfall, C.S.; Sherp, A.M.; Zubieta, C.; Alvarez, S.; Schraft, E.; Marcellin, R.; Ramirez, L.; Jez, J.M. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 13917–13922. [Google Scholar] [CrossRef]
- Makandar, R.; Nalam, V.J.; Chowdhury, Z.; Sarowar, S.; Klossner, G.; Lee, H.; Burdan, D.; Trick, H.N.; Gobbato, E.; Parker, J.E.; et al. The Combined Action of ENHANCED DISEASE SUSCEPTIBILITY1, PHYTOALEXIN DEFICIENT4, and SENESCENCE-ASSOCIATED101 Promotes Salicylic Acid-Mediated Defenses to Limit Fusarium graminearum Infection in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2015, 28, 943–953. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen Effectors Target Arabidopsis EDS1 and Alter Its Interactions with Immune Regulators. Science 2011, 334, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jeong, R.-D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 Forms a Ternary Complex with EDS1 and PAD4 and Is Required for Resistance Signaling against Turnip Crinkle Virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of Transcription Factor MYC2 by EDS1/PAD4 Complexes Bolsters Salicylic Acid Defense in Arabidopsis Effector-Triggered Immunity. Mol. Plant 2018, 11, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, S.; Suliman, M.; Bartsch, M.; Halder, V.; Maintz, J.; Bautor, J.; Zeier, J.; Parker, J.E.; Kombrink, E. Chemical Activation of EDS1/PAD4 Signaling Leading to Pathogen Resistance in Arabidopsis. Plant Cell Physiol. 2018, 59, 1592–1607. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z.; Sodmergen. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 2018, 69, 1051–1064. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Kwon, D.-J.; Jin, H.; Kang, M.-J.; Yun, S.-H.; Choi, S.-M.; Noh, Y.-S.; Noh, B. Salicylic acid-induced transcriptional reprogramming by the HAC–NPR1–TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 2018, 46, 11712–11725. [Google Scholar] [CrossRef]
- Krinke, O.; Flemr, M.; Vergnolle, C.; Collin, S.; Renou, J.-P.; Taconnat, L.; Yu, A.; Burketová, L.; Valentová, O.; Zachowski, A.; et al. Phospholipase D Activation Is an Early Component of the Salicylic Acid Signaling Pathway in Arabidopsis Cell Suspensions. Plant Physiol. 2009, 150, 424–436. [Google Scholar] [CrossRef]
- Krinke, O.; Ruelland, E.; Valentová, O.; Vergnolle, C.; Renou, J.-P.; Taconnat, L.; Flemr, M.; Burketová, L.; Zachowski, A. Phosphatidylinositol 4-Kinase Activation Is an Early Response to Salicylic Acid in Arabidopsis Suspension Cells. Plant Physiol. 2007, 144, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Janda, M.; Šašek, V.; Chmelařová, H.; Andrejch, J.; Nováková, M.; Hajšlová, J.; Burketová, L.; Valentová, O. Phospholipase D affects translocation of NPR1 to the nucleus in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Canet, J.V.; Dobon, A.; Roig, A.; Tornero, P. Structure-function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ. 2010, 33, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.; Garretón, V.; Frey, N.; Dominguez, C.; Pérez-Acle, T.; Van der Straeten, D.; Jordana, X.; Holuigue, L. Identification of NPR1-Dependent and Independent Genes Early Induced by Salicylic Acid Treatment in Arabidopsis. Plant Mol. Biol. 2005, 59, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Rong, D.; Luo, N.; Mollet, J.C.; Liu, X.; Yang, Z. Salicylic Acid Regulates Pollen Tip Growth through an NPR3/NPR4-Independent Pathway. Mol. Plant. 2016, 9, 1478–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelló, M.J.; Medina-Puche, L.; Lamilla, J.; Tornero, P. NPR1 paralogs of Arabidopsis and their role in salicylic acid perception. PLoS ONE 2018, 13, e0209835. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.; Tian, M.; Klessig, D.F. Salicylic acid binds NPR3 and NPR4 to regulate NPR1-dependent defense responses. Cell Res. 2012, 22, 1631. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreau, M.; Westlake, T.; Zampogna, G.; Popescu, G.; Tian, M.; Noutsos, C.; Popescu, S. The Arabidopsis oligopeptidases TOP1 and TOP2 are salicylic acid targets that modulate SA-mediated signaling and the immune response. Plant J. 2013, 76, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Tian, M.; Moreau, M.; Park, S.-W.; Choi, H.W.; Fei, Z.; Friso, G.; Asif, M.; Manosalva, P.; von Dahl, C.C.; et al. Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klessig, D.F.; Tian, M.; Choi, H.W. Multiple Targets of Salicylic Acid and Its Derivatives in Plants and Animals. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Klessig, D.F. Identification of a soluble salicylic acid-binding protein that may function in signal transduction in the plant disease-resistance response. Proc. Natl. Acad. Sci. USA 1991, 88, 8179–8183. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ricigliano, J.W.; Klessig, D.F. Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc. Natl. Acad. Sci. USA 1993, 90, 9533–9537. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Silva, H.; Klessig, D. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 1993, 262, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Chen, Z.; Ricigliano, J.R.; Klessig, D.F. Two inducers of plant defense responses, 2,6-dichloroisonicotinec acid and salicylic acid, inhibit catalase activity in tobacco. Proc. Natl. Acad. Sci. USA 1995, 92, 7143–7147. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-M.; Liu, W.-C.; Lu, Y.-T. CATALASE2 Coordinates SA-Mediated Repression of Both Auxin Accumulation and JA Biosynthesis in Plant Defenses. Cell Host Microbe 2017, 21, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Klessig, D.F. Identification of a Soluble, High-Affinity Salicylic Acid-Binding Protein in Tobacco. Plant Physiol. 1997, 113, 1319–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Klessig, D.F. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc. Natl. Acad. Sci. USA 2003, 100, 16101–16106. [Google Scholar] [CrossRef] [Green Version]
- Forouhar, F.; Yang, Y.; Kumar, D.; Chen, Y.; Fridman, E.; Park, S.W.; Chiang, Y.; Acton, T.B.; Montelione, G.T.; Pichersky, E.; et al. Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA 2005, 102, 1773–1778. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, D.; Jiang, Y.-L.; Kumar, D. SABP2, a methyl salicylate esterase is required for the systemic acquired resistance induced by acibenzolar-S-methyl in plants. FEBS Lett. 2010, 584, 3458–3463. [Google Scholar] [CrossRef]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469.e416. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Gustafsson, C.; Klessig, D.F. Validation of RNAi silencing specificity using synthetic genes: salicylic acid-binding protein 2 is required for innate immunity in plants. Plant J. 2006, 45, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-W.; Liu, P.-P.; Forouhar, F.; Vlot, A.C.; Tong, L.; Tietjen, K.; Klessig, D.F. Use of a synthetic salicylic acid analog to investigate the roles of methyl salicylate and its esterases in plant disease resistance. J. Biol. Chem. 2009, 284, 7307–7317. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Park, S.-W.; Forouhar, F.; Tong, L.; Fry, W.E.; Klessig, D.F. Methyl Esterase 1 (StMES1) Is Required for Systemic Acquired Resistance in Potato. Mol. Plant Microbe Interact. 2010, 23, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Guan, J.; Forouhar, F.; Tschaplinski, T.J.; Cheng, Z.-M.; Tong, L.; Chen, F. Two poplar methyl salicylate esterases display comparable biochemical properties but divergent expression patterns. Phytochemistry 2009, 70, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Liu, P.-P.; Cameron, R.K.; Park, S.-W.; Yang, Y.; Kumar, D.; Zhou, F.; Padukkavidana, T.; Gustafsson, C.; Pichersky, E.; et al. Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 2008, 56, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, D.H.; Navarre, D.A.; Clark, D.; del Pozo, O.; Martin, G.B.; Klessig, D.F. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 11640–11645. [Google Scholar] [CrossRef] [Green Version]
- DiMario, R.J.; Clayton, H.; Mukherjee, A.; Ludwig, M.; Moroney, J.V. Plant Carbonic Anhydrases: Structures, Locations, Evolution, and Physiological Roles. Mol. Plant 2017, 10, 30–46. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Feechan, A.; Yun, B.-W.; Shafiei, R.; Hofmann, A.; Taylor, P.; Xue, P.; Yang, F.-Q.; Xie, Z.-S.; Pallas, J.A.; et al. S-Nitrosylation of AtSABP3 Antagonizes the Expression of Plant Immunity. J. Biol. Chem. 2009, 284, 2131–2137. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, B.; Jaleel, H.; Sadiq, Y.; Khan, M.M.A.; Shabbir, A. Response of exogenous salicylic acid on cadmium induced photosynthetic damage, antioxidant metabolism and essential oil production in peppermint. Plant Growth Regul. 2018, 86, 273–286. [Google Scholar] [CrossRef]
- Medina-Puche, L.; Castelló, M.J.; Canet, J.V.; Lamilla, J.; Colombo, M.L.; Tornero, P. β-carbonic anhydrases play a role in salicylic acid perception in Arabidopsis. PLoS ONE 2017, 12, e0181820. [Google Scholar] [CrossRef] [PubMed]
- Poque, S.; Wu, H.-W.; Huang, C.-H.; Cheng, H.-W.; Hu, W.-C.; Yang, J.-Y.; Wang, D.; Yeh, S.-D. Potyviral gene-silencing suppressor HCPro interacts with salicylic acid (SA)-binding protein 3 to weaken SA-mediated defense responses. Mol. Plant Microbe Interact. 2017, 31, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Ruelland, E.; Miginiac-Maslow, M. Regulation of chloroplast enzyme activities by thioredoxins: Activation or relief from inhibition? Trends Plant Sci. 1999, 4, 136–141. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454.e1415–1467.e1415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant. J. 2006, 48, 647–656. [Google Scholar] [CrossRef]
- Liu, G.; Holub, E.B.; Alonso, J.M.; Ecker, J.R.; Fobert, P.R. An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant. J. 2005, 41, 304–318. [Google Scholar] [CrossRef]
- Chang, M.; Zhao, J.; Chen, H.; Li, G.; Chen, J.; Li, M.; Palmer, I.A.; Song, J.; Alfano, J.R.; Liu, F.; et al. PBS3 Protects EDS1 from Proteasome-Mediated Degradation in Plant Immunity. Mol. Plant 2019, 12, 678–688. [Google Scholar] [CrossRef]
- Zhou, X.-T.; Jia, L.-J.; Wang, H.-Y.; Zhao, P.; Wang, W.-Y.; Liu, N.; Song, S.-W.; Wu, Y.; Su, L.; Zhang, J.; et al. The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant. J. 2018, 95, 1055–1068. [Google Scholar] [CrossRef]
- Tian, M.; von Dahl, C.C.; Liu, P.-P.; Friso, G.; van Wijk, K.J.; Klessig, D.F. The combined use of photoaffinity labeling and surface plasmon resonance-based technology identifies multiple salicylic acid-binding proteins. Plant J. 2012, 72, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural Basis for Prereceptor Modulation of Plant Hormones by GH3 Proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef]
- Okrent, R.A.; Brooks, M.D.; Wildermuth, M.C. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 2009, 284, 9742–9754. [Google Scholar] [CrossRef] [PubMed]
- Schuman, M.C.; Meldau, S.; Gaquerel, E.; Diezel, C.; McGale, E.; Greenfield, S.; Baldwin, I.T. The Active Jasmonate JA-Ile Regulates a Specific Subset of Plant Jasmonate-Mediated Resistance to Herbivores in Nature. Front. Plant Sci. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobuta, K.; Okrent, R.A.; Stoutemyer, M.; Rodibaugh, N.; Kempema, L.; Wildermuth, M.C.; Innes, R.W. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007, 144, 1144–1156. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Sasvari, Z.; Gonzalez, P.A.; Friso, G.; Rowland, E.; Liu, X.-M.; van Wijk, K.J.; Nagy, P.D.; Klessig, D.F. Salicylic Acid Inhibits the Replication of Tomato bushy stunt virus by Directly Targeting a Host Component in the Replication Complex. Mol. Plant Microbe Interact. 2015, 28, 379–386. [Google Scholar] [CrossRef]
- Choi, H.W.; Tian, M.; Manohar, M.; Harraz, M.M.; Park, S.-W.; Schroeder, F.C.; Snyder, S.H.; Klessig, D.F. Human GAPDH Is a Target of Aspirin’s Primary Metabolite Salicylic Acid and Its Derivatives. PLoS ONE 2015, 10, e0143447. [Google Scholar] [CrossRef]
- Liao, Y.; Tian, M.; Zhang, H.; Li, X.; Wang, Y.; Xia, X.; Zhou, J.; Zhou, Y.; Yu, J.; Shi, K.; et al. Salicylic acid binding of mitochondrial alpha-ketoglutarate dehydrogenase E2 affects mitochondrial oxidative phosphorylation and electron transport chain components and plays a role in basal defense against tobacco mosaic virus in tomato. New Phytol. 2015, 205, 1296–1307. [Google Scholar] [CrossRef]
- Wang, R.; Rajagopalan, K.; Sadre-Bazzaz, K.; Moreau, M.; Klessig, D.F.; Tong, L. Structure of the Arabidopsis thaliana TOP2 oligopeptidase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 555–559. [Google Scholar] [CrossRef]
- Westlake, T.J.; Ricci, W.A.; Popescu, G.V.; Popescu, S.C. Dimerization and thiol sensitivity of the salicylic acid binding thimet oligopeptidases TOP1 and TOP2 define their functions in redox-sensitive cellular pathways. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Moissiard, G.; Bischof, S.; Husmann, D.; Pastor, W.A.; Hale, C.J.; Yen, L.; Stroud, H.; Papikian, A.; Vashisht, A.A.; Wohlschlegel, J.A.; et al. Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc. Natl. Acad. Sci. USA 2014, 111, 7474–7479. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Choi, H.W.; Manosalva, P.; Austin, C.A.; Peters, J.E.; Klessig, D.F. Plant and Human MORC Proteins Have DNA-Modifying Activities Similar to Type II Topoisomerases, but Require One or More Additional Factors for Full Activity. Mol. Plant Microbe Interact. 2017, 30, 87–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.W.; Manohar, M.; Manosalva, P.; Tian, M.; Moreau, M.; Klessig, D.F. Activation of Plant Innate Immunity by Extracellular High Mobility Group Box 3 and Its Inhibition by Salicylic Acid. PLoS Pathog. 2016, 12, e1005518. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinform. 2008, 2008, 5. [Google Scholar] [CrossRef] [PubMed]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genom. 2005, 21, 389–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Gupta, V.; Liu, S.; Ando, H.; Ishii, R.; Tateno, S.; Kaneko, Y.; Yugami, M.; Sakamoto, S.; Yamaguchi, Y.; Nureki, O.; et al. Salicylic Acid Induces Mitochondrial Injury by Inhibiting Ferrochelatase Heme Biosynthesis Activity. Mol. Pharmacol. 2013, 84, 824–833. [Google Scholar] [CrossRef] [Green Version]
- Delaney, T.P.; Friedrich, L.; Ryals, J.A. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 1995, 92, 6602–6606. [Google Scholar] [CrossRef]
- Belt, K.; Huang, S.; Thatcher, L.F.; Casarotto, H.; Singh, K.B.; Van Aken, O.; Millar, A.H. Salicylic Acid-Dependent Plant Stress Signaling via Mitochondrial Succinate Dehydrogenase. Plant Physiol. 2017, 173, 2029–2040. [Google Scholar] [CrossRef] [Green Version]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokotylo, I.; Kravets, V.; Ruelland, E. Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. Int. J. Mol. Sci. 2019, 20, 4377. https://doi.org/10.3390/ijms20184377
Pokotylo I, Kravets V, Ruelland E. Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. International Journal of Molecular Sciences. 2019; 20(18):4377. https://doi.org/10.3390/ijms20184377
Chicago/Turabian StylePokotylo, Igor, Volodymyr Kravets, and Eric Ruelland. 2019. "Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling" International Journal of Molecular Sciences 20, no. 18: 4377. https://doi.org/10.3390/ijms20184377
APA StylePokotylo, I., Kravets, V., & Ruelland, E. (2019). Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. International Journal of Molecular Sciences, 20(18), 4377. https://doi.org/10.3390/ijms20184377