Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants
Abstract
:1. Introduction
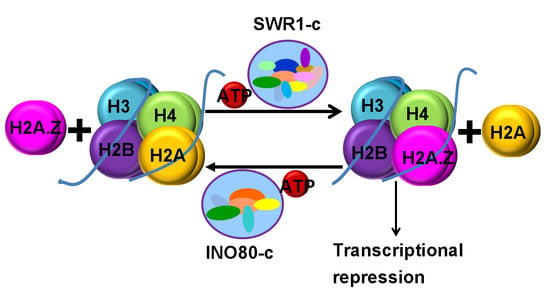
2. Chromatin Remodeling Functions of INO80 and SWR1 CRCs
3. The Subunits of INO80 and SWR1 CRCs
4. Involvement of INO80/SWR1-c in DNA Repair
5. Functions of INO80/SWR1-c in Flowering
6. Functions of INO80/SWR1-c in Immunity Response
7. Involvement of INO80/SWR1-c in the Regulation of MicroRNA Expression
8. Other Functions of Core Subunits of INO80/SWR1-c
9. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Act1 | Actin1 |
| ARP4 | Actin-Related Protein 4 |
| ARP5 | Actin-Related Protein 5 |
| ARP6 | Actin-Related Protein 6 |
| ARP8 | Actin-Related Protein 8 |
| ARP9 | Actin-Related Protein 9 |
| ATC | anaplastic thyroid carcinoma |
| ATP | adenosine triphosphate |
| ATPases | adenosine triphosphatase |
| Bdf1 | Bromodomain factor 1 |
| Brg1 | brahma-related gene 1 |
| CCA1 | CIRCADIAN CLOCK ASSOCIATED1 |
| CHD | chromo and DNA-binding domain |
| CRCs | chromatin-remodeling complexes |
| CSCsCUCs | cancer stem cellsCUP SHAPED COTYLEDONs |
| DMC1 | DISRUPTED MEIOTIC cDNA1 |
| DNA | DeoxyriboNucleic Acid |
| DSBs | DNA double-strand breaks |
| EAF1 | Esa1-associated factor 1 |
| ERF9 | ethylene response factor 9 |
| Esa1 | Essential for SAS Family Acetyltransferase 1 |
| ESD1 | EARLY IN SHORT DAYS 1 |
| FLC | FLOWERING LOCUS C |
| FLX | FLC EXPRESSOR |
| FT | FLOWERING LOCUS T |
| FUL | FRUITFULL |
| GI | GIGANTEA |
| HAM1 | HAIRY MERISTEM 1 |
| Hda1 | Histone deacetylase 1 |
| HR | homologous recombination |
| HSA | helicase/SANT-associated |
| hta11 | HISTONE H2A 11 |
| hta9 | HISTONE H2A 9 |
| IAA19 | AUXIN-RESPONSIVE PROTEIN 19 |
| Ies1-6 | Ino Eighty Subunit 1-6 |
| INO80 | Inositol Requiring 80 |
| INO80-c | INO80 complex |
| ISWI | imitation SWI |
| MAF4/5 | MADS AFFECTING FLOWERING 4/5 |
| MBD9 | Methyl-CpG-binding domain 9 |
| miRNAs | MicroRNAs |
| MMC | megaspore mother cells |
| NHEJ | nonhomologous end-joining |
| Nhp10 | Non-Histone Protein 10 |
| NRP1 | NAP1-RELATED PROTEIN 1 |
| NuA4 | nucleosome acetyltransferase of H4 |
| NuA4-c | NuA4 complex |
| PHD | plant homeodomain |
| PIE1 | PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 |
| PSR | phosphate starvation responses |
| RAD51/RAD54 | RADiation sensitive 51/54 |
| RPM1 | RESISTANCE TO P. SYRINGAE PV MACULICOLA 1 |
| RPP5 | RECOGNITION OF PERONOSPORA PARASITICA 5 |
| Rvb1 | RuvB-like protein 1 |
| RVB1/RIN1 | RESISTANCE TO PSEUDOMONAS SYRINGAE PV MACULICOLA INTERACTOR 1 |
| Rvb2 | RuvB-like protein 2 |
| RVB2A/B | RuvB-like protein 2A/B |
| SAGA | Spt-Ada-Gcn5-acetyltransferase |
| SANT | Swi3-Ada2-NCoR-TFIIIB |
| SAR | systemic acquired resistance |
| SEF | SERRATED LEAVES AND EARLY FLOWERING |
| SF2 | superfamily 2 |
| SHL 3 | superhelical location 3 |
| SHL2 | superhelical location 2 |
| SHL6 | superhelical location 6 |
| SKIP | SNW/SKI-INTERACTING PROTEIN |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| SPLs | SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-DOMAIN TRANSCRIPTION FACTORs |
| SRCAP | SNF2-RELATED CBP ACTIVATOR PROTEIN (human) |
| SUF3 | SUPPRESSOR OF FRI 3 |
| SUF4 | SUPPRESSOR OF FRI 4 |
| SVP | SHORT VEGETATIVE PHASE |
| Swc2-6 | SWR complex subunit 2-6 |
| SWI | mating type switching |
| SWR1 | SWi2/snf2-Related 1 |
| SWR1-c | SWR1 complex |
| Taf14 | TATA binding protein-Associated Factor 14 |
| TAP-MS | tandem mass spectrometry |
| TCPs | TEOSINTE BRANCHED 1, CYCLOIDEA AND PCF TRANSCRIPTION FACTORs |
| Tip60 | Tat-interactive protein 60 |
| Tra1 | transfer region 1 |
| TRRAP | transformation/transcription domain-associated protein |
| Yaf9 | YEAST ALL1-FUSED GENE FROM CHROMOSOME 9 |
| YAF9A | YEAST ALL1-FUSED GENE FROM CHROMOSOME 9A |
| YAF9A/B | YEAST ALL1-FUSED GENE FROM CHROMOSOME 9A/B |
| YAF9B | YEAST ALL1-FUSED GENE FROM CHROMOSOME 9B |
References
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Shilatifard, A. Histone modification: Cause or cog? Trends Genet. 2011, 27, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Jerzmanowski, A. Swi/snf chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 2007, 1769, 330–345. [Google Scholar] [CrossRef] [PubMed]
- Imbalzano, A.N. Energy-dependent chromatin remodelers: Complex complexes and their components. Crit. Rev. Eukar. Gene 1998, 8, 225–255. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Koonin, E.V. Helicases-amino-acid-sequence comparisons and structure-function-relationships. Curr. Opin. Struc. Biol. 1993, 3, 419–429. [Google Scholar] [CrossRef]
- Abrams, E.; Neigeborn, L.; Carlson, M. Molecular analysis of snf2 and snf5, genes required for expression of glucose-repressible genes in saccharomyces-cerevisiae. Mol. Cell Biol. 1986, 6, 3643–3651. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Morrison, A.J.; Shen, X.T. Chromatin remodelling beyond transcription: The ino80 and swr1 complexes. Nat. Rev. Mol. Cell Biol. 2009, 10, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Gerhold, C.B.; Gasser, S.M. Ino80 and swr complexes: Relating structure to function in chromatin remodeling. Trends Cell Biol. 2014, 24, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. Atp-driven exchange of histone h2az variant catalyzed by swr1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global regulation of h2a.Z localization by the ino80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Tan, D.; Lakshminarasimhan, M.; Washburn, M.P.; Hong, E.J.; Walz, T.; Peterson, C.L. Structural analyses of the chromatin remodelling enzymes ino80-c and swr-c. Nat. Commun. 2015, 6, 7108. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Radman-Livaja, M.; Rando, O.J.; Peterson, C.L. A histone acetylation switch regulates h2a.Z deposition by the swr-c remodeling enzyme. Science 2013, 340, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ranjan, A.; Wei, D.B.; Wu, C. Comment on “a histone acetylation switch regulates h2a.Z deposition by the swr-c remodeling enzyme”. Science 2016, 353, 358. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Peterson, C.L. Response to comment on “a histone acetylation switch regulates h2a.Z deposition by the swr-c remodeling enzyme”. Science 2016, 353, 358. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.; Schall, K.; Kostrewa, D.; Lakomek, K.; Strauss, M.; Moldt, M.; Hopfner, K.P. Structural basis for atp-dependent chromatin remodelling by the ino80 complex. Nature 2018, 556, 386–390. [Google Scholar] [CrossRef]
- Willhoft, O.; Ghoneim, M.; Lin, C.L.; Chua, E.Y.D.; Wilkinson, M.; Chaban, Y.; Ayala, R.; McCormack, E.A.; Ocloo, L.; Rueda, D.S.; et al. Structure and dynamics of the yeast swr1-nucleosome complex. Science 2018, 362, eaat7716. [Google Scholar] [CrossRef]
- Li, B.; Pattenden, S.G.; Lee, D.; Gutierrez, J.; Chen, J.; Seidel, C.; Gerton, J.; Workman, J.L. Preferential occupancy of histone variant h2az at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 2005, 102, 18385–18390. [Google Scholar] [CrossRef]
- Yen, K.; Vinayachandran, V.; Pugh, B.F. Swr-c and ino80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 2013, 154, 1246–1256. [Google Scholar] [CrossRef]
- Zilberman, D.; Coleman-Derr, D.; Ballinger, T.; Henikoff, S. Histone h2a.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 2008, 456, 125–129. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Zilberman, D. Deposition of histone variant h2a.Z within gene bodies regulates responsive genes. PLoS Genet. 2012, 8, e1002988. [Google Scholar] [CrossRef] [PubMed]
- Ebbert, R.; Birkmann, A.; Schuller, H.J. The product of the snf2/swi2 paralogue ino80 of saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 1999, 32, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Mizuguchi, G.; Hamiche, A.; Wu, C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 2000, 406, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.H.; Wu, C.H.; Ladurner, A.; Mizuguchi, G.; Wei, D.; Xiao, H.; Luk, E.; Ranjan, A.; Wu, C. N terminus of swr1 binds to histone h2az and provides a platform for subunit assembly in the chromatin remodeling complex. J. Biol. Chem. 2009, 284, 6200–6207. [Google Scholar] [CrossRef] [PubMed]
- Allard, S.; Utley, R.T.; Savard, J.; Clarke, A.; Grant, P.; Brandl, C.J.; Pillus, L.; Workman, J.L.; Cote, J. Nua4, an essential transcription adaptor/histone h4 acetyltransferase complex containing esa1p and the atm-related cofactor tra1p. Embo J. 1999, 18, 5108–5119. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Cote, J. The highly conserved and multifunctional nua4 hat complex. Curr. Opin. Genet. Dev. 2004, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.; Lambert, J.P.; Gerdes, M.; Al-Madhoun, A.S.; Skerjanc, I.S.; Figeys, D.; Baetz, K. Functional dissection of the nua4 histone acetyltransferase reveals its role as a genetic hub and that eaf1 is essential for complex integrity. Mol. Cell Biol. 2008, 28, 2244–2256. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.; Auger, A.; Monnet-Saksouk, J.; Brodeur, J.; Piquet, S.; Cramet, M.; Bouchard, N.; Lacoste, N.; Utley, R.T.; Gaudreau, L.; et al. Nua4-dependent acetylation of nucleosomal histones h4 and h2a directly stimulates incorporation of h2a.Z by the swr1 complex. J. Biol. Chem. 2010, 285, 15966–15977. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Halley, J.E.; Rine, J. Telomeric heterochromatin boundaries require nua4-dependent acetylation of histone variant h2a.Z in saccharomyces cerevisiae. Gene Dev. 2006, 20, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.Y.; Lu, J.Y.; Zhang, J.M.; Walter, W.; Dang, W.W.; Wan, J.; Tao, S.C.; Qian, J.; Zhao, Y.M.; Boeke, J.D.; et al. Protein acetylation microarray reveals that nua4 controls key metabolic target regulating gluconeogenesis. Cell 2009, 136, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Kamine, J.; Elangovan, B.; Subramanian, T.; Coleman, D.; Chinnadurai, G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the hiv-1 tat transactivator. Virology 1996, 216, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Ikura, T.; Ogryzko, V.V.; Grigoriev, M.; Groisman, R.; Wang, J.; Horikoshi, M.; Scully, R.; Qin, J.; Nakatani, Y. Involvement of the tip60 histone acetylase complex in DNA repair and apoptosis. Cell 2000, 102, 463–473. [Google Scholar] [CrossRef]
- Legube, G.; Linares, L.K.; Lemercier, C.; Scheffner, M.; Khochbin, S.; Trouche, D. Tip60 is targeted to proteasome-mediated degradation by mdm2 and accumulates after uv irradiation. Embo J. 2002, 21, 1704–1712. [Google Scholar] [CrossRef] [PubMed]
- Giaimo, B.D.; Ferrante, F.; Vallejo, D.M.; Hein, K.; Gutierrez-Perez, I.; Nist, A.; Stiewe, T.; Mittler, G.; Herold, S.; Zimmermann, T.; et al. Histone variant h2a.Z deposition and acetylation directs the canonical notch signaling response. Nucleic Acids Res. 2018, 46, 8197–8215. [Google Scholar] [CrossRef] [PubMed]
- Auger, A.; Galarneau, L.; Altaf, M.; Nourani, A.; Doyon, Y.; Utley, R.T.; Cronier, D.; Allard, S.; Cote, J. Eaf1 is the platform for nua4 molecular assembly that evolutionarily links chromatin acetylation to atp-dependent exchange of histone h2a variants. Mol. Cell Biol. 2008, 28, 2257–2270. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, W.; Chang, P.; Wu, H.; Liu, H.; Chen, J. Merge and separation of nua4 and swr1 complexes control cell fate plasticity in candida albicans. Cell Discov. 2018, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.S.; Amasino, R.M. Pie1, an iswi family gene, is required for flc activation and floral repression in arabidopsis. Plant Cell 2003, 15, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Zambrano, Á.; Crevillén, P.; Franco-Zorrilla, J.M.; López, J.A.; Santos-González, J.; Roszak, P.; Santos-Gonzalez, J.; Jurado, S.; Vázquez, J.; Köhler, C.; et al. Arabidopsis swc4 binds DNA and recruits the swr1 complex to modulate histone h2a.Z deposition at key regulatory genes. Mol. Plant 2018, 11, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Sijacic, P.; Holder, D.H.; Bajic, M.; Deal, R.B. Methyl-cpg-binding domain 9 (mbd9) is required for h2a.Z incorporation into chromatin at a subset of h2a.Z-enriched regions in the arabidopsis genome. PLoS Genet. 2019, 15, e1008326. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, O.; Benvenuto, G.; Bowler, C.; Molinier, J.; Hohn, B. The ino80 protein controls homologous recombination in arabidopsis thaliana. Mol. Cell 2004, 16, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Crevillén, P.; Gómez-Zambrano, Á.; López, J.A.; Vázquez, J.; Piñeiro, M.; Jarillo, J.A. Arabidopsis yaf9 histone readers modulate flowering time through nua4-complex-dependent h4 and h2a.Z histone acetylation at flc chromatin. NEW Phytol. 2019, 222, 1893–1908. [Google Scholar] [CrossRef] [PubMed]
- Van Attikum, H.; Fritsch, O.; Hohn, B.; Gasser, S.M. Recruitment of the ino80 complex by h2a phosphorylation links atp-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004, 119, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.J.; Highland, J.; Krogan, N.J.; Arbel-Eden, A.; Greenblatt, J.F.; Haber, J.E.; Shen, X. Ino80 and gamma-h2ax interaction links atp-dependent chromatin remodeling to DNA damage repair. Cell 2004, 119, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Van Attikum, H.; Fritsch, O.; Gasser, S.M. Distinct roles for swr1 and ino80 chromatin remodeling complexes at chromosomal double-strand breaks. Embo J. 2007, 26, 4113–4125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cao, L.; Rong, L.; An, Z.; Zhou, W.; Ma, J.; Shen, W.H.; Zhu, Y.; Dong, A. The chromatin-remodeling factor atino80 plays crucial roles in genome stability maintenance and in plant development. Plant J. 2015, 82, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Gao, J.; Ma, J.; Cao, L.; Zhang, C.; Zhu, Y.; Dong, A.; Shen, W.H. Distinct roles of the histone chaperones nap1 and nrp and the chromatin-remodeling factor ino80 in somatic homologous recombination in arabidopsis thaliana. Plant J. 2016, 88, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Ma, J.; Wu, D.; Shen, W.H.; Zhu, Y. Functional coordination of the chromatin-remodeling factor atino80 and the histone chaperones nrp1/2 in inflorescence meristem and root apical meristem. Front. Plant Sci. 2019, 10, 115. [Google Scholar] [CrossRef]
- Muller, J.; Oma, Y.; Vallar, L.; Friederich, E.; Poch, O.; Winsor, B. Sequence and comparative genomic analysis of actin-related proteins. Mol. Biol. Cell 2005, 16, 5736–5748. [Google Scholar] [CrossRef] [PubMed]
- Szerlong, H.; Hinata, K.; Viswanathan, R.; Erdjument-Bromage, H.; Tempst, P.; Cairns, B.R. The hsa domain binds nuclear actin-related proteins to regulate chromatin-remodeling atpases. Nat. Struct. Mol. Biol. 2008, 15, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; McKinney, E.C.; Deal, R.B.; Smith, A.P.; Meagher, R.B. Arabidopsis actin-related protein arp5 in multicellular development and DNA repair. Dev. Biol. 2009, 335, 22–32. [Google Scholar] [CrossRef]
- Kang, H.; Zhang, C.; An, Z.; Shen, W.H.; Zhu, Y. Atino80 and atarp5 physically interact and play common as well as distinct roles in regulating plant growth and development. NEW Phytol. 2019, 223, 336–353. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, J.; Hwang, H.J.; Kim, S.; Park, C.; Kim, S.Y.; Lee, I. The frigida complex activates transcription of flc, a strong flowering repressor in arabidopsis, by recruiting chromatin modification factors. Plant Cell 2011, 23, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, A.; Gómez-Zambrano, A.; López-González, L.; Piñeiro, M.; Jarillo, J.A. Mutations in the arabidopsis swc6 gene, encoding a component of the swr1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 2008, 59, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, J.; Muller, S.Y.; Oh, M.; Underwood, C.; Henderson, I.; Lee, I. Regulation of microrna-mediated developmental changes by the swr1 chromatin remodeling complex. Plant Physiol. 2016, 171, 1128–1143. [Google Scholar] [PubMed]
- Xu, M.L.; Leichty, A.R.; Hu, T.Q.; Poethig, R.S. H2a.Z promotes the transcription of mir156a and mir156c in arabidopsis by facilitating the deposition of h3k4me3. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Bieluszewski, T.; Galganski, L.; Sura, W.; Bieluszewska, A.; Abram, M.; Ludwikow, A.; Ziolkowski, P.A.; Sadowski, J. Ateaf1 is a potential platform protein for arabidopsis nua4 acetyltransferase complex. BMC Plant Biol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Berriri, S.; Gangappa, S.N.; Kumar, S.V. Swr1 chromatin-remodeling complex subunits and h2a.Z have non-overlapping functions in immunity and gene regulation in arabidopsis. Mol. Plant 2016, 9, 1051–1065. [Google Scholar] [CrossRef] [PubMed]
- Holt, B.F.; Boyes, D.C.; Ellerstrom, M.; Siefers, N.; Wiig, A.; Kauffman, S.; Grant, M.R.; Dangl, J.L. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal arabidopsis development. Dev. Cell 2002, 2, 807–817. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Silencing the nuclear actin-related protein atarp4 in arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 2005, 41, 845–858. [Google Scholar] [CrossRef]
- March-Díaz, R.; García-Domínguez, M.; Florencio, F.J.; Reyes, J.C. Sef, a new protein required for flowering repression in arabidopsis, interacts with pie1 and arp6. Plant Physiol. 2007, 143, 893–901. [Google Scholar] [CrossRef]
- Choi, K.; Park, C.; Lee, J.; Oh, M.; Noh, B.; Lee, I. Arabidopsis homologs of components of the swr1 complex regulate flowering and plant development. Development 2007, 134, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Deal, R.B.; Topp, C.N.; McKinney, E.C.; Meagher, R.B. Repression of flowering in arabidopsis requires activation of flowering locus c expression by the histone variant h2a.Z. Plant Cell 2007, 19, 74–83. [Google Scholar] [CrossRef] [PubMed]
- March-Díaz, R.; García-Domínguez, M.; Lozano-Juste, J.; León, J.; Florencio, F.J.; Reyes, J.C. Histone h2a.Z and homologues of components of the swr1 complex are required to control immunity in arabidopsis. Plant J. 2008, 53, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, M.; Chai, M.; He, Q.; Huang, X.; Zhao, L.; Qin, Y. Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between h2a.Z and h3k4me3. New Phytol. 2019, 221, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.; Bishop, B.; Ho, K.K.; Huang, R.; Jia, W.; Zhang, H.; Pascuzzi, P.E.; Deal, R.B.; Ogas, J. The chromatin remodelers pkl and pie1 act in an epigenetic pathway that determines h3k27me3 homeostasis in arabidopsis. Plant Cell 2018, 30, 1337–1352. [Google Scholar] [CrossRef]
- Choi, K.; Kim, S.; Kim, S.Y.; Kim, M.; Hyun, Y.; Lee, H.; Choe, S.; Kim, S.G.; Michaels, S.; Lee, I. Suppressor of frigida3 encodes a nuclear actin-related protein6 required for floral repression in arabidopsis. Plant Cell 2005, 17, 2647–2660. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Lazaro, A.; Poethig, R.S.; Gomez-Mena, C.; Pineiro, M.A.; Martinez-Zapater, J.M.; Jarillo, J.A. Early in short days 1 (esd1) encodes actin-related protein 6 (atarp6), a putative component of chromatin remodelling complexes that positively regulates flc accumulation in arabidopsis. Development 2006, 133, 1241–1252. [Google Scholar] [CrossRef]
- Rosa, M.; Von Harder, M.; Cigliano, R.A.; Schlogelhofer, P.; Mittelsten Scheid, O. The arabidopsis swr1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 2013, 25, 1990–2001. [Google Scholar] [CrossRef]
- Qin, Y.; Zhao, L.H.; Skaggs, M.I.; Andreuzza, S.; Tsukamoto, T.; Panoli, A.; Wallace, K.N.; Smith, S.; Siddiqi, I.; Yang, Z.B.; et al. Actin-related protein6 regulates female meiosis by modulating meiotic gene expression in arabidopsis. Plant Cell 2014, 26, 1612–1628. [Google Scholar] [CrossRef]
- Zhao, L.; Cai, H.; Su, Z.; Wang, L.; Huang, X.; Zhang, M.; Chen, P.; Dai, X.; Zhao, H.; Palanivelu, R.; et al. Klu suppresses megasporocyte cell fate through swr1-mediated activation of wrky28 expression in arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E526–E535. [Google Scholar] [CrossRef]
- Sura, W.; Kabza, M.; Karlowski, W.M.; Bieluszewski, T.; Kus-Slowinska, M.; Paweloszek, L.; Sadowski, J.; Ziolkowski, P.A. Dual role of the histone variant h2a.Z in transcriptional regulation of stress-response genes. Plant Cell 2017, 29, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P.; Jain, A.; Deal, R.B.; Nagarajan, V.K.; Poling, M.D.; Raghothama, K.G.; Meagher, R.B. Histone h2a.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 2010, 152, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Zacharaki, V.; Benhamed, M.; Poulios, S.; Latrasse, D.; Papoutsoglou, P.; Delarue, M.; Vlachonasios, K.E. The arabidopsis ortholog of the yeats domain containing protein yaf9a regulates flowering by controlling h4 acetylation levels at the flc locus. Plant Sci. 2012, 196, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Wang, S.L.; Zhang, F.; Zheng, H.; Liu, Y.N.; Huang, T.T.; Ding, Y. Phosphorylation of histone h2a at serine 95: A plant-specific mark involved in flowering time regulation and h2a.Z deposition. Plant Cell 2017, 29, 2197–2213. [Google Scholar] [CrossRef] [PubMed]
- Wils, C.R.; Kaufmann, K. Gene-regulatory networks controlling inflorescence and flower development in arabidopsis thaliana. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of svp in the control of flowering time by ambient temperature in arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Laurie, R.; Macknight, R. It’s time to flower: The genetic control of flowering time. Bioessays 2004, 26, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Kobor, M.S.; Venkatasubrahmanyam, S.; Meneghini, M.D.; Gin, J.W.; Jennings, J.L.; Link, A.J.; Madhani, H.D.; Rine, J. A protein complex containing the conserved swi2/snf2-related atpase swr1p deposits histone variant h2a.Z into euchromatin. Plos Biol. 2004, 2, 587–599. [Google Scholar] [CrossRef]
- Cui, Z.; Tong, A.; Huo, Y.; Yan, Z.; Yang, W.; Yang, X.; Wang, X.X. Skip controls flowering time via the alternative splicing of sef pre-mrna in arabidopsis. BMC Biol. 2017, 15, 80. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, H.; Xing, L.; Xu, S.; Liu, H.; Chong, K.; Xu, Y. Requirement of histone acetyltransferases ham1 and ham2 for epigenetic modification of flc in regulating flowering in arabidopsis. J. Plant Physiol. 2013, 170, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Le Masson, I.; Yu, D.Y.; Jensen, K.; Chevalier, A.; Courbeyrette, R.; Boulard, Y.; Smith, M.M.; Mann, C. Yaf9, a novel nua4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell Biol. 2003, 23, 6086–6102. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. Yeats domain proteins: A diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 2009, 87, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.J.; Ahmad, S.; Vann, K.R.; Andrews, F.H.; Mayo, Z.A.; Bourriquen, G.; Bridgers, J.B.; Zhang, J.Y.; Strahl, B.D.; Cote, J.; et al. Yaf9 subunit of the nua4 and swr1 complexes targets histone h3k27ac through its yeats domain. Nucleic Acids Res. 2018, 46, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Richardson, D.O.; Roberts, D.N.; Utley, R.; Erdjument-Bromage, H.; Tempst, P.; Cote, J.; Cairns, B.R. The yaf9 component of the swr1 and nua4 complexes is required for proper gene expression, histone h4 acetylation, and htz1 replacement near telomeres. Mol. Cell Biol. 2004, 24, 9424–9436. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.Y.T.; Levesque, N.; Kobor, M.S. Nua4 and swr1-c: Two chromatin-modifying complexes with overlapping functions and components. Biochem Cell Biol. 2009, 87, 799–815. [Google Scholar] [CrossRef] [PubMed]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. Aaa+: A class of chaperone-like atpases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [PubMed]
- Kanemaki, M.; Kurokawa, Y.; Matsu-ura, T.; Makino, Y.; Masani, A.; Okazaki, K.; Morishita, T.; Tamura, T. Tip49b, a new ruvb-like DNA helicase, is included in a complex together with another ruvb-like DNA helicase, tip49a. J. Biol. Chem. 1999, 274, 22437–22444. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary conservation of microrna regulatory programs in plant flower development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. Microrna and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, Y.; Gong, Y.; Dong, T.; Zhang, B.; Gao, W. Mir-148a inhibits self-renewal of thyroid cancer stem cells via repressing ino80 expression. Oncol. Rep. 2016, 36, 3387–3396. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Bai, Y.; Zhao, L.; Dou, X.; Liu, Y.; Wang, L.; Li, Y.; Li, W.; Hui, Y.; Huang, X.; et al. H2a.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in arabidopsis. Mol. Plant 2017, 10, 1274–1292. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.; Park, C.; Choi, K.; Lee, D.; Seok, C.; Lee, I. Molecular evolution of actin related protein 6, a component of swr1 complex in arabidopsis. J. Plant Biol. 2016, 59, 467–477. [Google Scholar] [CrossRef]
- Cheng, X.; Auger, A.; Altaf, M.; Drouin, S.; Paquet, E.; Utley, R.T.; Robert, F.; Cote, J. Eaf1 links the nua4 histone acetyltransferase complex to htz1 incorporation and regulation of purine biosynthesis. Eukaryot Cell 2015, 14, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Ladurner, A.G.; Inouye, C.; Jain, R.; Tjian, R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 2003, 11, 365–376. [Google Scholar] [CrossRef]
- Durant, M.; Pugh, B.F. Nua4-directed chromatin transactions throughout the saccharomyces cerevisiae genome. Mol. Cell Biol. 2007, 27, 5327–5335. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J. Lysine acetylation and the bromodomain: A new partnership for signaling. Bioessays 2004, 26, 1076–1087. [Google Scholar] [CrossRef]

| Complex | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Organism | Yeast | Human | Arabidopsis | ||||||
| Family and Composition | INO80 | SWR1 | NuA4 | INO80 | SWR1/SRCAP | NuA4/Tip60 | INO80 | SWR1 | NuA4 |
| ATPase or Acetyltransferase | Ino80 | Swr1 | Eaf1 **, Esa1 * | hIno80 | SRCAP | p400 **, Tip60 * | INO80 | PIE1 | HAM1/2 *, EAF1 ** |
| Noncatalytic homologous subunits | Rvb1, Rvb2 | Tip49a, Tip49b | RVB1/RIN1 | ||||||
| RVB2A, RVB2B | |||||||||
| Arp4, Actin1 | BAF53a | ARP4, ACT1 | |||||||
| Arp5, Arp8 | Arp6 | Arp5, Arp8 | Arp6 | Actin | ARP5, ARP9 | ARP6 | |||
| Taf14 | Yaf9 | GAS41 | GAS41/YAF9A, TAF14/YAF9B | ||||||
| Ies2, Ies6 | hIes2, hIes6 | ||||||||
| Swc4/Eaf2 | DMAP1 | SWC4 | |||||||
| Swc2/Vps72 | YL-1 | SWC2 | |||||||
| Swc6/Vps71 | ZnF-HIT1 | SWC6 | |||||||
| Bdf1 | Brd8/TRCp120 | ||||||||
| H2A.Z, H2B | H2A.Z, H2B | H2A.Z, H2B | |||||||
| Tra1 | TRRAP | TRA1 | |||||||
| Epl1 | EPC1 | ||||||||
| Yng2 | ING3 | ING1, ING2 | |||||||
| Eaf3 | MRG15 | MRG1, MRG2 | |||||||
| Eaf5 | |||||||||
| Eaf7 | MRGBP | ||||||||
| Eaf6 | hEaf6 | ||||||||
| Unique | Ies1, Ies3, Ies 4, Ies5, Nhp10 | Swc3,5,7 | Amida, NFRKB, MCRS1, FLJ90652, FLJ20309 | MRGX, FLJ11730, MRGBP, EPC1, EPC-like | MBD9, AL5-7 | ||||
| Gene | Interacting Proteins * | Functions | Reference | |
|---|---|---|---|---|
| The common subunits of INO80/SWR1-c | SWC2 | unknown | ||
| SWC4 | Flowering time control; Leaf cell proliferation and expansion | [38] | ||
| SWC6/SEF | SUF4, FLX, TAF14 | Flowering time control | [52,53] | |
| MicroRNA expression | [54,55] | |||
| HAM1, EAF1 | [38,56] | |||
| Immunity response | [57] | |||
| RVB1/RIN1 | FLX, SUF4, FES1, FRI | [52] | ||
| RPM1, RPP5 | Sporophyte and female gametophyte; Disease resistance | [58] | ||
| RVB2A, RVB2B | EAF1 | [56] | ||
| ARP4 | Multiple effects on plant development | [59] | ||
| Core subunit of SWR1, NuA4, INO80 and SWI/SNF complexes | [56] | |||
| Unique subunits of INO80-c | INO80 | Controls homologous recombination | [40,45,46,51] | |
| Flowering time control | [45,51] | |||
| Apical meristems maintenance | [47] | |||
| ARP5 | organ development; DNA repair | [50,51] | ||
| ARP9 | unknown | |||
| Unique subunits of SWR1-c | PIE1 | Flowering time control | [37,60,61,62] | |
| Immunity response | [57,63] | |||
| MicroRNA expression | [54] | |||
| Anthocyanin biosynthesis | [64] | |||
| Maintenance of H3K27me3 | [65] | |||
| ARP6 | Flowering time control | [61,62,66,67] | ||
| MicroRNA expression | [54,55] | |||
| Immunity response | [57] | |||
| DNA repair | [68] | |||
| Female meiosis regulation; Germ-line specification | [69,70] | |||
| Osmotic stress; Phosphate starvation response | [71,72] | |||
| YAF9A | CCA1, HAM1 | Flowering time control | [41,73,74] | |
| YAF9B |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Gao, S.; Peng, X.; Wu, K.; Yang, S. Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants. Int. J. Mol. Sci. 2019, 20, 4591. https://doi.org/10.3390/ijms20184591
Wang J, Gao S, Peng X, Wu K, Yang S. Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants. International Journal of Molecular Sciences. 2019; 20(18):4591. https://doi.org/10.3390/ijms20184591
Chicago/Turabian StyleWang, Jianhao, Sujuan Gao, Xiuling Peng, Keqiang Wu, and Songguang Yang. 2019. "Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants" International Journal of Molecular Sciences 20, no. 18: 4591. https://doi.org/10.3390/ijms20184591
APA StyleWang, J., Gao, S., Peng, X., Wu, K., & Yang, S. (2019). Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants. International Journal of Molecular Sciences, 20(18), 4591. https://doi.org/10.3390/ijms20184591