Sequence and Structural Analysis of AA9 and AA10 LPMOs: An Insight into the Basis of Substrate Specificity and Regioselectivity
Abstract
:1. Introduction
2. Results
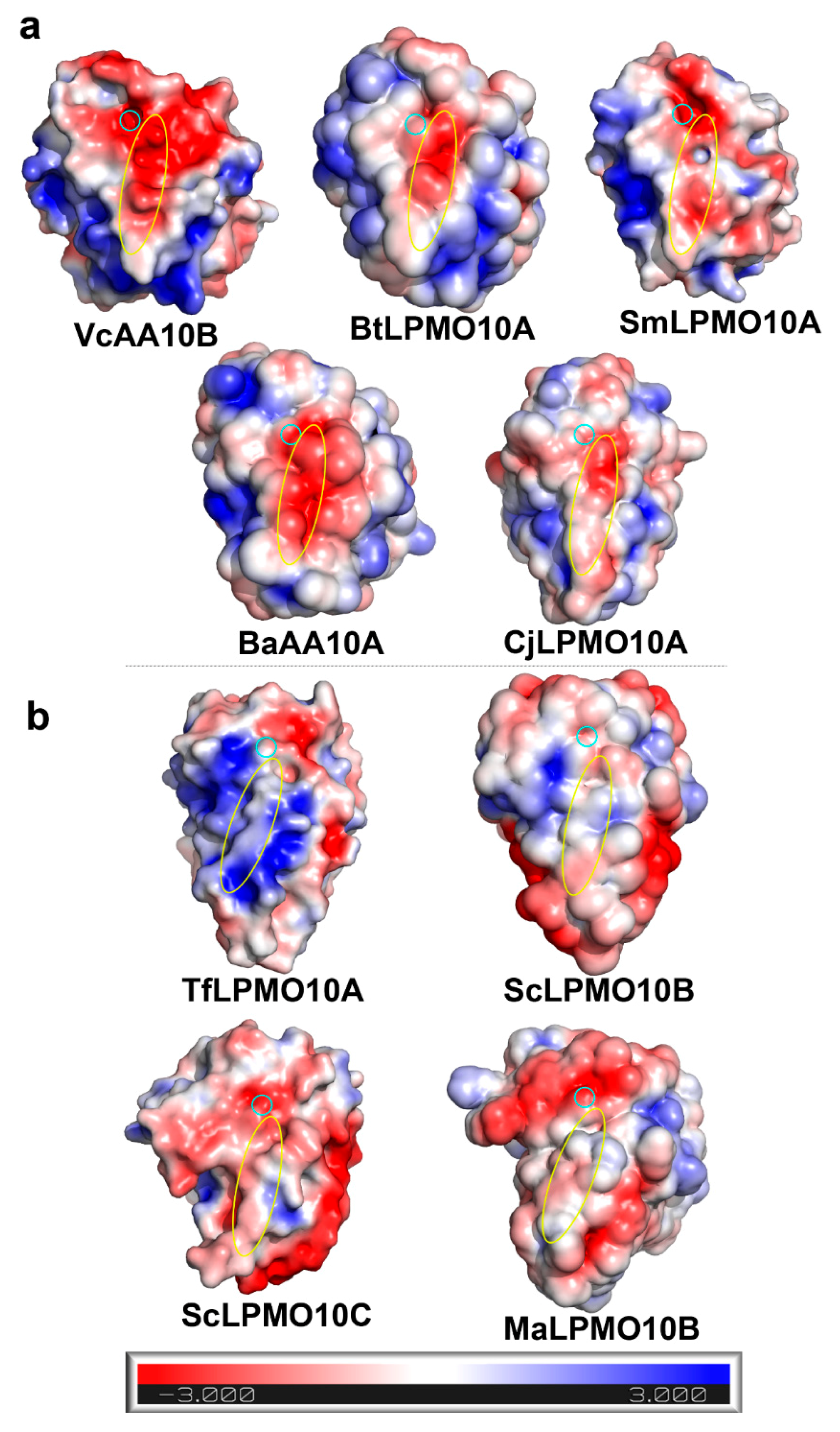
2.1. A Motif on L2 Loop Affects the Substrate Specificity of AA10 LPMOs
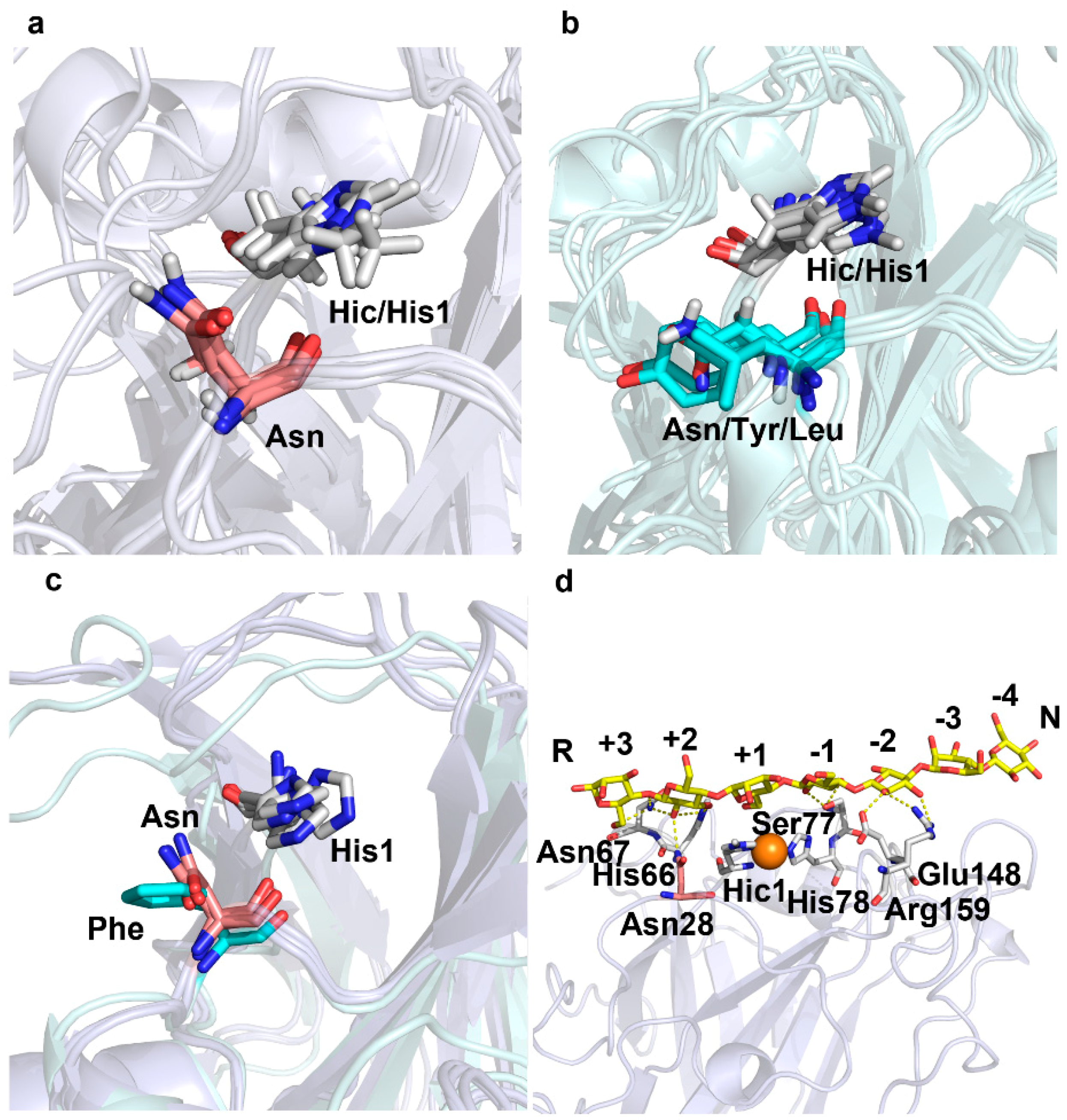
2.2. A Conserved Asparagine at 7Å of Catalytic Cu Contributes to C4-Oxidation
2.3. Appended Modules Are Related to Substrate Specificity but Not Regioselectivity
3. Discussion
4. Conclusions
5. Materials and Methods
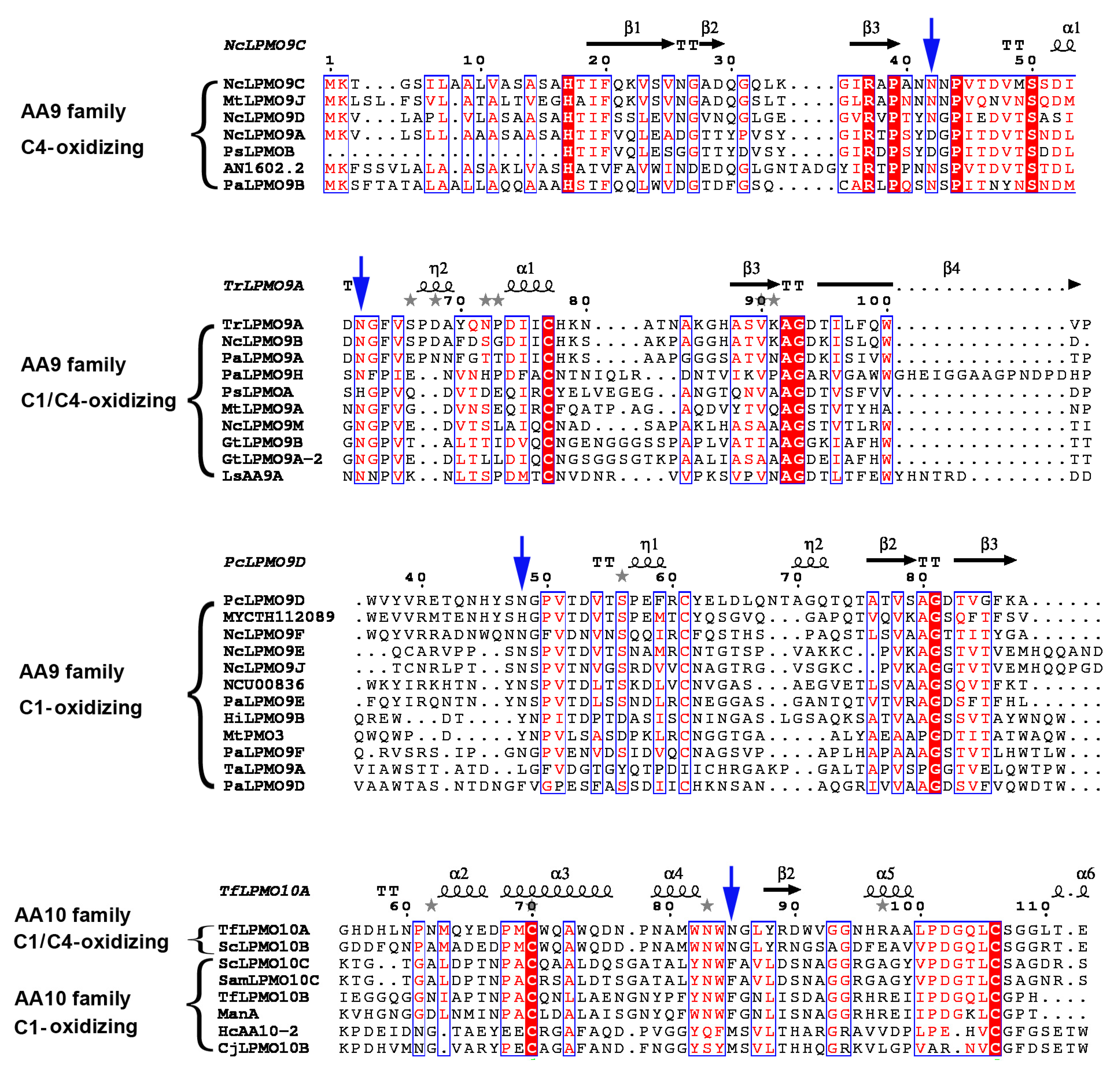
5.1. Multiple Sequence Alignment and Phylogenetic Tree Construction
5.2. Motif Identification
5.3. Electrostatic Potential and pKa Value Calculation
5.4. Protein Structure Analysis and Molecular Docking
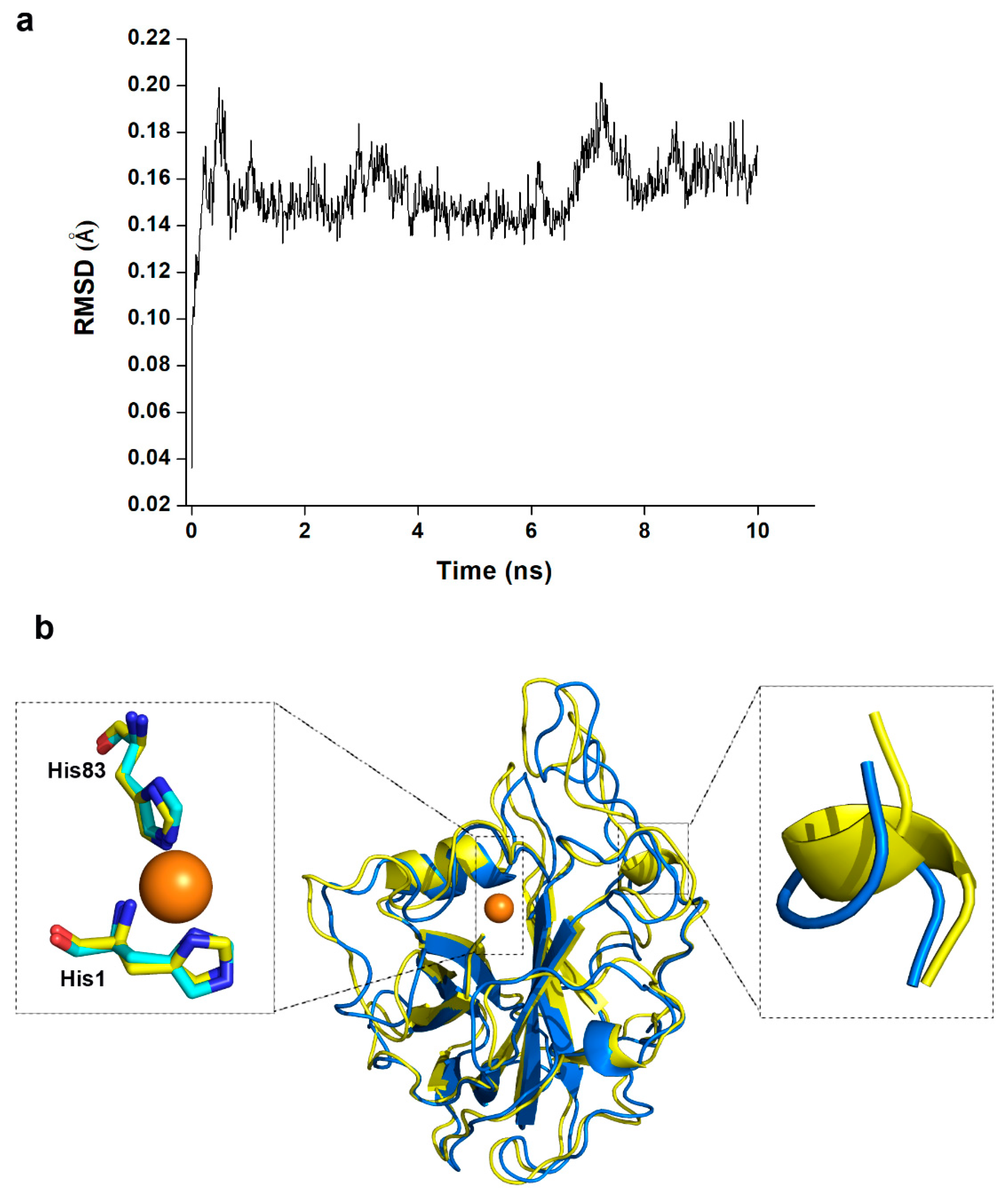
5.5. Homology Modelling and Molecular Dynamic Simulation
5.6. Sequence Similarity Network
Author Contributions
Funding
Conflicts of Interest
References
- Vaaje-Kolstad, G.W.B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Sorlie, M.; Petrović, D.; Courtade, G.; Aachmann, F.L.; Vaaje-Kolstad, G.; Bissaro, B.; Røhr, Å.K.; Eijsink, V.G. Polysaccharide degradation by lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 2019, 59, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Johansen, K.S. Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 2016, 44, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Meier, K.K.; Jones, S.M.; Kaper, T.; Hansson, H.; Koetsier, M.J.; Karkehabadi, S.; Solomon, E.I.; Sandgren, M.; Kelemen, B. Oxygen Activation by Cu LPMOs in Recalcitrant Carbohydrate Polysaccharide Conversion to Monomer Sugars. Chem. Rev. 2018, 118, 2593–2635. [Google Scholar] [CrossRef] [PubMed]
- Tandrup, T.; Frandsen, K.E.H.; Johansen, K.S.; Berrin, J.G.; Lo Leggio, L. Recent insights into lytic polysaccharide monooxygenases (LPMOs). Biochem. Soc. Trans. 2018, 46, 1431–1447. [Google Scholar] [CrossRef]
- Filiatrault-Chastel, C.; Navarro, D.; Haon, M.; Grisel, S.; Herpoel-Gimbert, I.; Chevret, D.; Fanuel, M.; Henrissat, B.; Heiss-Blanquet, S.; Margeot, A.; et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol. Biofuels 2019, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Correa, T.L.; dos Santos, L.V.; Pereira, G.A. AA9 and AA10: From enigmatic to essential enzymes. Appl. Microbiol. Biotechnol. 2016, 100, 9–16. [Google Scholar] [CrossRef]
- Book, A.J.; Yennamalli, R.M.; Takasuka, T.E.; Currie, C.R.; Phillips, G.N., Jr.; Fox, B.G. Evolution of substrate specificity in bacterial AA10 lytic polysaccharide monooxygenases. Biotechnol. Biofuels 2014, 7, 109. [Google Scholar] [CrossRef] [Green Version]
- Hemsworth, G.R.; Henrissat, B.; Davies, G.J.; Walton, P.H. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat. Chem. Biol. 2014, 10, 122–126. [Google Scholar] [CrossRef]
- Vu, V.V.; Beeson, W.T.; Span, E.A.; Farquhar, E.R.; Marletta, M.A. A family of starch-active polysaccharide monooxygenases. Proc. Natl. Acad. Sci. USA 2014, 111, 13822–13827. [Google Scholar] [CrossRef] [Green Version]
- Couturier, M.; Ladeveze, S.; Sulzenbacher, G.; Ciano, L.; Fanuel, M.; Moreau, C.; Villares, A.; Cathala, B.; Chaspoul, F.; Frandsen, K.E.; et al. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat. Chem. Biol. 2018, 14, 306–310. [Google Scholar] [CrossRef] [Green Version]
- Sabbadin, F.; Hemsworth, G.R.; Ciano, L.; Henrissat, B.; Dupree, P.; Tryfona, T.; Marques, R.D.S.; Sweeney, S.T.; Besser, K.; Elias, L.; et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 2018, 9, 756. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Forsberg, Z.; Loose, J.S.; Bissaro, B.; Eijsink, V.G. Structural diversity of lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 2017, 44, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Courtade, G.; Forsberg, Z.; Heggset, E.B.; Eijsink, V.G.H.; Aachmann, F.L. The carbohydrate-binding module and linker of a modular lytic polysaccharide monooxygenase promote localized cellulose oxidation. J. Biol. Chem. 2018, 293, 13006–13015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansson, H.; Karkehabadi, S.; Mikkelsen, N.; Douglas, N.R.; Kim, S.; Lam, A.; Kaper, T.; Kelemen, B.; Meier, K.K.; Jones, S.M.; et al. High-resolution structure of a lytic polysaccharide monooxygenase from Hypocrea jecorina reveals a predicted linker as an integral part of the catalytic domain. J. Biol. Chem. 2017, 292, 19099–19109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crouch, L.I.; Labourel, A.; Walton, P.H.; Davies, G.J.; Gilbert, H.J. The Contribution of Non-catalytic Carbohydrate Binding Modules to the Activity of Lytic Polysaccharide Monooxygenases. J. Biol. Chem. 2016, 291, 7439–7449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruer-Zerhusen, N.; Alahuhta, M.; Lunin, V.V.; Himmel, M.E.; Bomble, Y.J.; Wilson, D.B. Structure of a Thermobifida fusca lytic polysaccharide monooxygenase and mutagenesis of key residues. Biotechnol. Biofuels 2017, 10, 243. [Google Scholar] [CrossRef]
- Span, E.A.; Marletta, M.A. The framework of polysaccharide monooxygenase structure and chemistry. Curr. Opin. Struct. Biol. 2015, 35, 93–99. [Google Scholar] [CrossRef]
- Li, X.; Beeson, W.T.t.; Phillips, C.M.; Marletta, M.A.; Cate, J.H. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 2012, 20, 1051–1061. [Google Scholar] [CrossRef]
- Frommhagen, M.; Koetsier, M.J.; Westphal, A.H.; Visser, J.; Hinz, S.W.; Vincken, J.P.; van Berkel, W.J.; Kabel, M.A.; Gruppen, H. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol. Biofuels 2016, 9, 186. [Google Scholar] [CrossRef]
- Frandsen, K.E.; Simmons, T.J.; Dupree, P.; Poulsen, J.C.; Hemsworth, G.R.; Ciano, L.; Johnston, E.M.; Tovborg, M.; Johansen, K.S.; von Freiesleben, P.; et al. The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat. Chem. Biol. 2016, 12, 298–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, T.J.; Frandsen, K.E.H.; Ciano, L.; Tryfona, T.; Lenfant, N.; Poulsen, J.C.; Wilson, L.F.L.; Tandrup, T.; Tovborg, M.; Schnorr, K.; et al. Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates. Nat. Commun. 2017, 8, 1064. [Google Scholar] [CrossRef] [PubMed]
- Bissaro, B.; Isaksen, I.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Rohr, A.K. How a Lytic Polysaccharide Monooxygenase Binds Crystalline Chitin. Biochemistry 2018, 57, 1893–1906. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Kognole, A.A.; Wu, M.; Westereng, B.; Crowley, M.F.; Kim, S.; Dimarogona, M.; Payne, C.M.; Sandgren, M. Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition. FEBS J. 2018, 285, 2225–2242. [Google Scholar] [CrossRef] [PubMed]
- Vu, V.V.; Beeson, W.T.; Phillips, C.M.; Cate, J.H.; Marletta, M.A. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J. Am. Chem. Soc. 2014, 136, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Beeson, W.T.; Vu, V.V.; Span, E.A.; Phillips, C.M.; Marletta, M.A. Cellulose degradation by polysaccharide monooxygenases. Annu Rev. Biochem. 2015, 84, 923–946. [Google Scholar] [CrossRef] [PubMed]
- Frommhagen, M.; Westphal, A.H.; van Berkel, W.J.H.; Kabel, M.A. Distinct Substrate Specificities and Electron-Donating Systems of Fungal Lytic Polysaccharide Monooxygenases. Front. Microbiol. 2018, 9, 1080. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Forsberg, Z.; Nelson, C.E.; Dalhus, B.; Mekasha, S.; Loose, J.S.; Crouch, L.I.; Rohr, A.K.; Gardner, J.G.; Eijsink, V.G.; Vaaje-Kolstad, G. Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio japonicus. J. Biol. Chem. 2016, 291, 7300–7312. [Google Scholar] [CrossRef] [Green Version]
- Wong, E.; Vaaje-Kolstad, G.; Ghosh, A.; Hurtado-Guerrero, R.; Konarev, P.V.; Ibrahim, A.F.; Svergun, D.I.; Eijsink, V.G.; Chatterjee, N.S.; van Aalten, D.M. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 2012, 8, e1002373. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Houston, D.R.; Riemen, A.H.; Eijsink, V.G.; van Aalten, D.M. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 2005, 280, 11313–11319. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Taylor, E.J.; Kim, R.Q.; Gregory, R.C.; Lewis, S.J.; Turkenburg, J.P.; Parkin, A.; Davies, G.J.; Walton, P.H. The copper active site of CBM33 polysaccharide oxygenases. J. Am. Chem. Soc. 2013, 135, 6069–6077. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Mackenzie, A.K.; Sorlie, M.; Rohr, A.K.; Helland, R.; Arvai, A.S.; Vaaje-Kolstad, G.; Eijsink, V.G. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc. Natl. Acad. Sci. USA 2014, 111, 8446–8451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsberg, Z.; Bissaro, B.; Gullesen, J.; Dalhus, B.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J. Biol. Chem. 2018, 293, 1397–1412. [Google Scholar] [CrossRef] [Green Version]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Borisova, A.S.; Isaksen, T.; Dimarogona, M.; Kognole, A.A.; Mathiesen, G.; Varnai, A.; Rohr, A.K.; Payne, C.M.; Sorlie, M.; Sandgren, M.; et al. Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity. J. Biol. Chem. 2015, 290, 22955–22969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, R.J.; Sweeney, M.D.; Lo Leggio, L.; Otten, H.; Poulsen, J.C.; Johansen, K.S.; Krogh, K.B.; Jorgensen, C.I.; Tovborg, M.; Anthonsen, A.; et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 2011, 108, 15079–15084. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Beckham, G.T.; Larsson, A.M.; Ishida, T.; Kim, S.; Payne, C.M.; Himmel, M.E.; Crowley, M.F.; Horn, S.J.; Westereng, B.; et al. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the Basidiomycota fungus Phanerochaete chrysosporium. J. Biol. Chem. 2013, 288, 12828–12839. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.C.; Kracher, D.; Gandini, R.; Sygmund, C.; Kittl, R.; Haltrich, D.; Hallberg, B.M.; Ludwig, R.; Divne, C. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat. Commun. 2015, 6, 7542. [Google Scholar] [CrossRef] [Green Version]
- Span, E.A.; Suess, D.L.M.; Deller, M.C.; Britt, R.D.; Marletta, M.A. The Role of the Secondary Coordination Sphere in a Fungal Polysaccharide Monooxygenase. ACS Chem. Biol. 2017, 12, 1095–1103. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Danneels, B.; Tanghe, M.; Desmet, T. Structural Features on the Substrate-Binding Surface of Fungal Lytic Polysaccharide Monooxygenases Determine Their Oxidative Regioselectivity. Biotechnol. J. 2019, 14, e1800211. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]



| Complex | Polar Contacts | Distance 1 (Å) a | Distance 2 (Å) b |
|---|---|---|---|
| NcLPMO9C–G7 | BMA322:O3-Asn26:ND2 BMA322:O2-Glu65:OE2 BGC324:O3-Ser82:OG BMA325:O2-His155:O BMA325:O3-His155:O BMA325:O2-Tyr204:OH BMA325:O3- Tyr204:OH BMA325:O6- Tyr204:OH | 5.0 | 4.0 |
| NcLPMO9C_N26F–G7 | BMA320:O6- Glu65:OE2 BGC323:O3-Ser82:OG BGC324:O2-Asn156:OD1 BGC324:O3-Asn156:OD1 BGC324:O3-His155:O BMA325:O4-Asn202:ND2 | 4.4 | 5.1 |
| TaLPMO9A–G7 | BGC326:O3-Asn20:O BGC324:O2-HIC:O BMA322:O2-Ser85:OG BMA322:O3-Ser85:OG BMA322:O6-Ser165:OG BGC321:O3- Ser165:OG BGC321:O2-Gln167:NE2 | 4.2 | 5.6 |
| Residue | pKa | Residue | pKa | ||
|---|---|---|---|---|---|
| WT | N26F | WT | N26F | ||
| Asp 13 | 2.59 | 2.59 | Tyr 90 | 13.49 | 13.49 |
| Asp 31 | 3.38 | 3.38 | Tyr 133 | 10.47 | 10.47 |
| Asp 36 | 4.35 | 4.35 | Tyr 145 | 12.01 | 11.98 |
| Asp 46 | 2.69 | 2.69 | Tyr 166 | 11.99 | 12.02 |
| Asp 74 | 3.19 | 3.19 | Tyr 192 | 18.6 | 18.59 |
| Asp 76 | 3.86 | 3.86 | Tyr 204 | 10.64 | 10.64 |
| Asp 95 | 3.23 | 3.23 | Tyr 217 | 14.13 | 14.13 |
| Asp 123 | 2.61 | 2.61 | Lys 6 | 11.06 | 11.06 |
| Asp 124 | 4.49 | 4.49 | Lys 18 | 10.43 | 10.43 |
| Asp 135 | 3.96 | 3.96 | Lys 45 | 9.58 | 9.58 |
| Asp 196 | 4.15 | 4.15 | Lys 57 | 10.55 | 10.55 |
| Asp 211 | 3.09 | 3.09 | Lys 84 | 10.77 | 10.77 |
| Glu 65 | 4.66 | 5.01 | Lys 93 | 9.55 | 9.55 |
| Glu 112 | 4.59 | 4.58 | Lys 106 | 10.38 | 10.38 |
| Glu 150 | 4.37 | 4.37 | Lys 109 | 9.54 | 9.54 |
| His 1 | 3.66 | 3.51 | Lys 119 | 10.55 | 10.55 |
| His 60 | 3.44 | 3.44 | Lys 209 | 10.5 | 10.5 |
| His 64 | 6.85 | 6.83 | Lys 215 | 10.2 | 10.2 |
| His 83 | 1.47 | 1.39 | Arg 21 | 10.95 | 10.95 |
| His 155 | 4.31 | 4.28 | Arg 148 | 14.86 | 14.86 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Qi, X.; Huang, H.; Zhu, H. Sequence and Structural Analysis of AA9 and AA10 LPMOs: An Insight into the Basis of Substrate Specificity and Regioselectivity. Int. J. Mol. Sci. 2019, 20, 4594. https://doi.org/10.3390/ijms20184594
Zhou X, Qi X, Huang H, Zhu H. Sequence and Structural Analysis of AA9 and AA10 LPMOs: An Insight into the Basis of Substrate Specificity and Regioselectivity. International Journal of Molecular Sciences. 2019; 20(18):4594. https://doi.org/10.3390/ijms20184594
Chicago/Turabian StyleZhou, Xiaoli, Xiaohua Qi, Hongxia Huang, and Honghui Zhu. 2019. "Sequence and Structural Analysis of AA9 and AA10 LPMOs: An Insight into the Basis of Substrate Specificity and Regioselectivity" International Journal of Molecular Sciences 20, no. 18: 4594. https://doi.org/10.3390/ijms20184594
APA StyleZhou, X., Qi, X., Huang, H., & Zhu, H. (2019). Sequence and Structural Analysis of AA9 and AA10 LPMOs: An Insight into the Basis of Substrate Specificity and Regioselectivity. International Journal of Molecular Sciences, 20(18), 4594. https://doi.org/10.3390/ijms20184594





