Overexpression of BpCUC2 Influences Leaf Shape and Internode Development in Betula pendula
Abstract
1. Introduction
2. Results
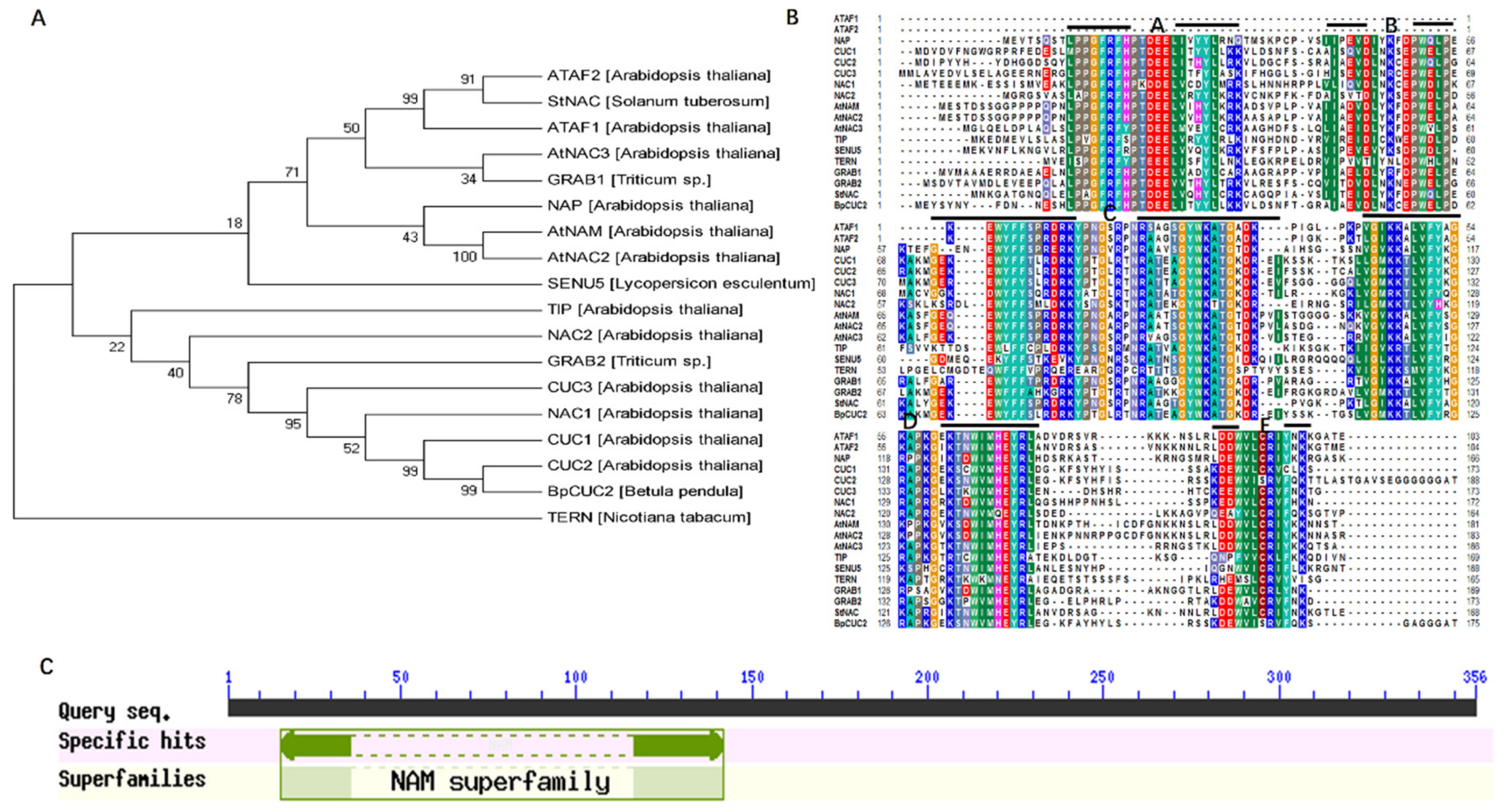
2.1. Identification of the CUC2 Gene in Betula pendula
2.2. Functional Analysis of BpCUC2 Gene
2.3. BpCUC2 Affects Epidermal Cell Size and Cell Number
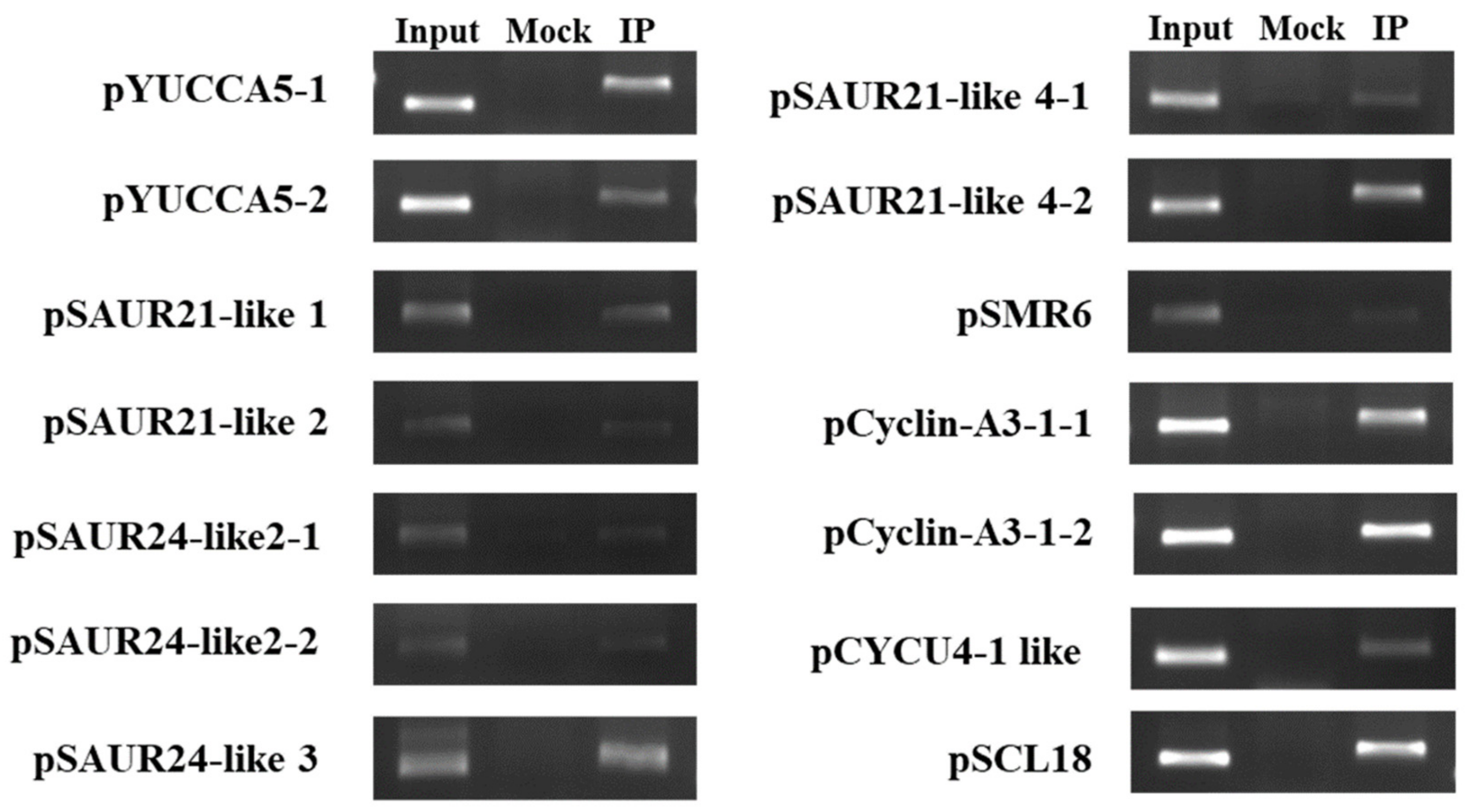
2.4. Identification of BpCUC2 Regulated Genes
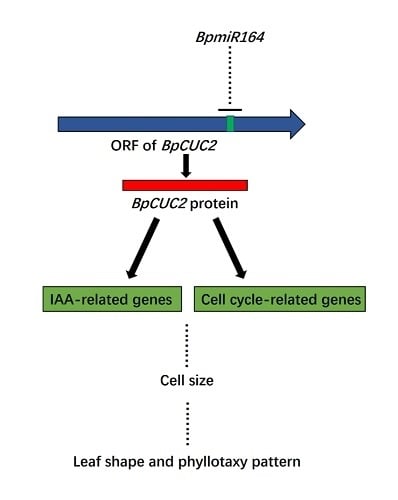
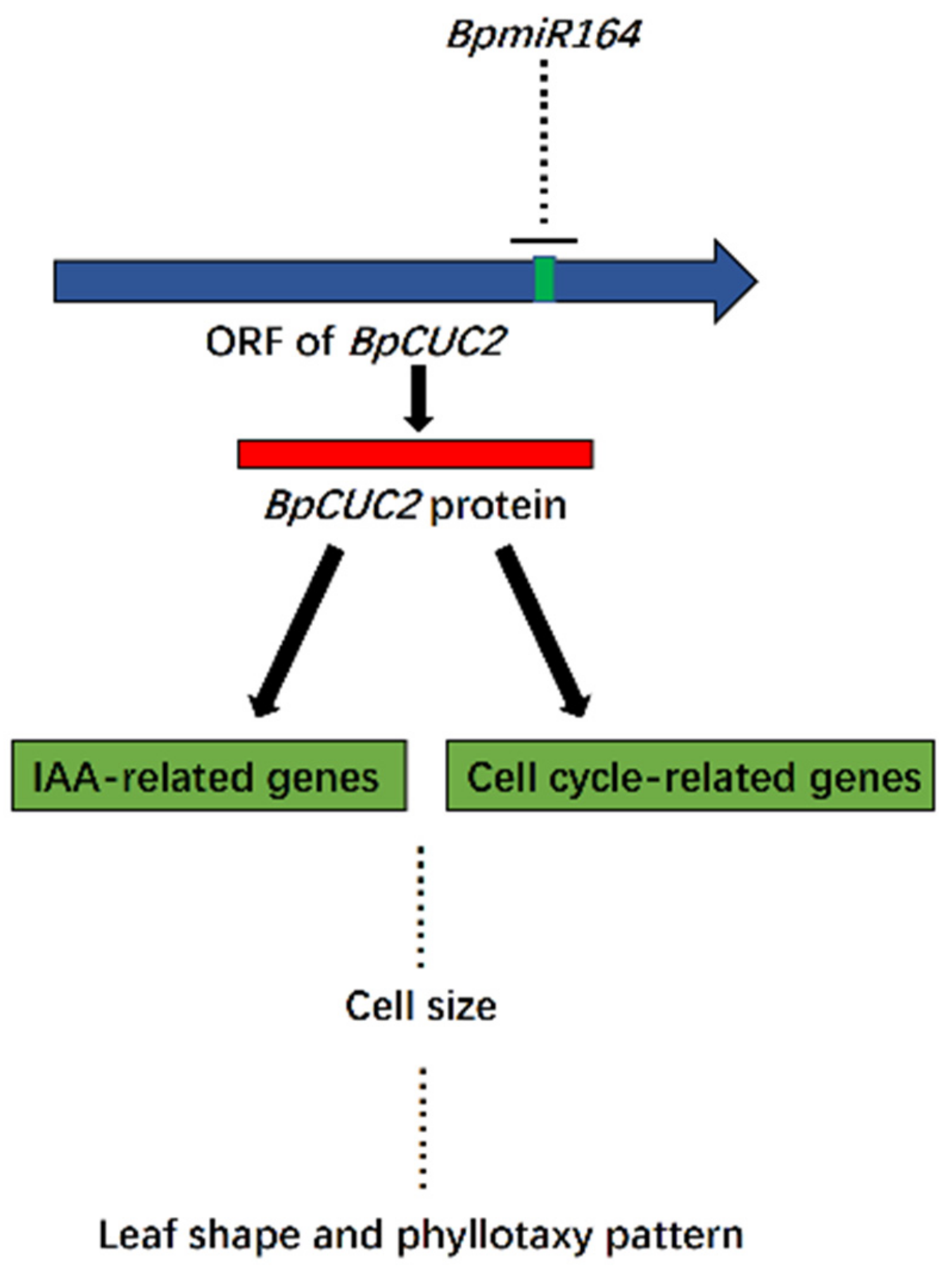
2.5. BpmiR164 Targets BpCUC2
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Identification and Bioinformatics Analysis of BpCUC2
4.3. Vector Construction and Birch Transformation
4.4. Analysis of Transformants
4.5. Analysis of BpCUC2 Expression
4.6. Internode Epidermal Cells Analysis
4.7. Histological Analyses and Microscopy
4.8. Phenotypic Characterization
4.9. Subcellular Localization
4.10. Transcriptome Analysis
4.11. Binding of BpCUC2 to Motifs
4.12. ChIP Assay
4.13. 5′ RACE
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ingram, G.C.; Waites, R. Keeping it together: Co-ordinating plant growth. Curr. Opin. Plant Biol. 2006, 9, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Tasaka, M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 1999, 126, 1563–1570. [Google Scholar] [PubMed]
- Takada, S.; Hibara, K.; Ishida, T.; Tasaka, M. The cup-shaped COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar] [PubMed]
- Xie, M.; Zhang, S.; Yu, B. microRNA biogenesis, degradation and activity in plants. Cell Mol. Life Sci. 2015, 72, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.C.; Sieber, P.; Wellmer, F.; Meyerowitz, E.M. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005, 15, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Wellmer, F.; Gheyselinck, J.; Riechmann, J.L.; Meyerowitz, E.M. Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 2007, 134, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Laufs, P.; Peaucelle, A.; Morin, H.; Traas, J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322. [Google Scholar] [CrossRef]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 Genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Peaucelle, A.; Morin, H.; Traas, J.; Laufs, P. Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 2007, 134, 1045–1050. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, D.; Liu, G.; Yang, C.; Teskey, R.O. Additive tree biomass equations for Betula platyphylla Suk. plantations in Northeast China. Ann. For. Sci. 2018, 75, 60. [Google Scholar] [CrossRef]
- Mu, H.; Lin, L.; Liu, G.; Jiang, J. Transcriptomic analysis of incised leaf-shape determination in birch. Gene 2013, 531, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Qu, C.; Zhang, M.; Li, Y.; Han, R.; Jiang, J.; Liu, G. Transcriptome sequencing to reveal the genetic regulation of leaf margin variation at early stage in birch. Tree Genet. Genomes 2018, 15, 4. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant J. 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Hibara, K.I.; Karim, M.R.; Takada, S.; Taoka, K.I.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef] [PubMed]
- Hasson, A.; Plessis, A.; Blein, T.; Adroher, B.; Grigg, S.; Tsiantis, M.; Boudaoud, A.; Damerval, C.; Laufs, P. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 2011, 23, 54–68. [Google Scholar] [CrossRef]
- Leyser, H.M.O. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 2014, 116, 397–403. [Google Scholar]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.Y.; Wang, L.Q.; Nie, X.G.; He, L.; Zang, D.D.; Liu, Y.J.; Zhang, B.; Wang, Y. A novel method to identify the DNA motifs recognized by a defined transcription factor. Plant Mol. Biol. 2014, 86, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.S.; Karrer, E.E.; Thomas, B.R.; Chen, L.; Rodriguez, R.L. Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 1998, 36, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Sutoh, K.; Yamauchi, D. Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 2003, 34, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Song, F.; Goodman, R.M.; Zheng, Z. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Biol. 2005, 7, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5 ’-region of Arabidopsis thaliana corl5a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Song, F.; Goodman, R.M.; Zheng, Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeo-domain transcriptional factor, in disease resistance responses. Plant Biol. 2005, 7, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.A.T.; Engler, J.D.A.; Raes, J.; Magyar, Z.; De Groodt, R.; Inzé, D.; Veylder, L. Molecular characterization of Arabidopsis PHO80-like proteins, a novel class of CDKA; 1-interacting cyclins. Cell. Mol. Life Sci. 2004, 61, 1485–1497. [Google Scholar]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and lateral suppressor controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Singh, D.; Tang, X. Essential RNA-based technologies and their applications in plant functional genomics. Trends Biotechnol. 2016, 34, 106–123. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, M.; Manrique, S.; Colombo, L.; Cuesta, C.; Benkova, E.; Novak, O. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. J. Exp. Bot. 2018, 69, 5169–5176. [Google Scholar] [CrossRef]
- Li, X.G.; Su, Y.H.; Zhao, X.Y.; Li, W.; Gao, X.Q.; Zhang, X.S. Cytokinin overproduction-caused alteration of flower development is partially mediated by CUC2 and CUC3 in Arabidopsis. Gene 2010, 450, 109–120. [Google Scholar] [CrossRef]
- Lee, D.-K.; Geisler, M.; Springer, P.S. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 2009, 136, 2423–2432. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, H.; Jiang, J.; Wang, S.; Liu, G. Analysis of the promoter features of BpCUC2 in Betula platyphylla × Betula pendula. Plant Cell. Tissue Organ Cult. 2018, 132, 191–199. [Google Scholar] [CrossRef]
- Hurst, C.H.; Turnbull, D.; Myles, S.M.; Leslie, K.; Keinath, N.F.; Hemsley, P.A. Variable Effects of C-Terminal Fusions on FLS2 Function: Not All Epitope Tags Are Created Equal. Plant Physiol. 2018, 177, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef]
- Marlene, R.; Anthony, A.M. Specificity of plant microRNA target MIMICs: corss-targeting of miR159 and miR319. Plant J. 2015, 180, 44–48. [Google Scholar]
- Velasquez, S.M.; Barbez, E.; Kleine-Vehn, J.; Estevez, J.M. Auxin and cellular elongation. Plant Physiol. 2016, 170, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Fedorova, E.; Lebris, M.; Lecomte, P.; Györgyey, J.; Vaubert, D.; Horvath, G.; Abad, P.; Kondorosi, A.; Kondorosi, E. The Medicago Species A2-Type cyclin is auxin regulated and involved in meristem formation but sispensable for endoreduplication-associated developmental programs. Plant Physiol. 2003, 131, 1091–1103. [Google Scholar] [CrossRef]
- Hartig, K.; Beck, E. Crosstalk between auxin, cytokinins, and sugars in the plant cell cycle. Plant Biol. 2006, 8, 389–396. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Wang, S.; Liu, G.; Li, H.; Huang, H.; Jiang, J. BpGH3.5, an early auxin-response gene, regulates root elongation in Betula platyphylla × Betula pendula. Plant Cell. Tissue Organ Cult. 2015, 120, 239–250. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evo. 1987, 4, 406–425. [Google Scholar]
- Marchler-bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; Deweese-scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shujun, C.; Jeff, P.; John, C. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 1993, 11, 113–116. [Google Scholar]
- Yang, L.; Li, Y.; Shen, H. Somatic embryogenesis and plant regeneration from immature zygotic embryo cultures of mountain ash (Sorbus pohuashanensis). Plant Cell. Tissue Organ Cult. 2012, 109, 547–556. [Google Scholar] [CrossRef]
- Ash, A.; Ellis, B.; Hickey, L.J.; Johnson, K.; Wilf, P.; Wing, S. Manual of Leaf Architecture Morphological Description and Categorization of Dicotyledonous and Net-Veined Monocotyledonous Angiosperms; Smithsonian Institution: Washington, DC, USA, 1999; ISBN 0967755409. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ji, X.; Zheng, L.; Liu, Y.; Nie, X. A Transient transformation system for the functional characterization of genes involved in stress response. Plant Mol. Biol. Report 2014, 32, 732–739. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C.; et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef]
- Haring, M.; Offermann, S.; Danker, T.; Horst, I.; Peterhansel, C.; Stam, M. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 2007, 16, 1–16. [Google Scholar] [CrossRef]







| Clone Number | Random DNA Insertion Sequence (5′-3′) with Two Sides of the Flanking Sequences | Motif Prediction |
|---|---|---|
| 3 | CCAGGCGTCGGG | CGACGOSAMY3 (CGACG), LTRECOREATCOR15 (CCGAC) |
| 1 | CAACTCCGAGTG | CAREOSREP1 (CAACTC) |
| 1 | CTGCTTGTCAGG | BIHD1OS (TGTCA), WRKY71OS motif (TGAC) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Xu, H.; Han, R.; Wang, S.; Liu, G.; Chen, S.; Chen, J.; Bian, X.; Jiang, J. Overexpression of BpCUC2 Influences Leaf Shape and Internode Development in Betula pendula. Int. J. Mol. Sci. 2019, 20, 4722. https://doi.org/10.3390/ijms20194722
Liu C, Xu H, Han R, Wang S, Liu G, Chen S, Chen J, Bian X, Jiang J. Overexpression of BpCUC2 Influences Leaf Shape and Internode Development in Betula pendula. International Journal of Molecular Sciences. 2019; 20(19):4722. https://doi.org/10.3390/ijms20194722
Chicago/Turabian StyleLiu, Chaoyi, Huanwen Xu, Rui Han, Shuo Wang, Guifeng Liu, Su Chen, Jiying Chen, Xiuyan Bian, and Jing Jiang. 2019. "Overexpression of BpCUC2 Influences Leaf Shape and Internode Development in Betula pendula" International Journal of Molecular Sciences 20, no. 19: 4722. https://doi.org/10.3390/ijms20194722
APA StyleLiu, C., Xu, H., Han, R., Wang, S., Liu, G., Chen, S., Chen, J., Bian, X., & Jiang, J. (2019). Overexpression of BpCUC2 Influences Leaf Shape and Internode Development in Betula pendula. International Journal of Molecular Sciences, 20(19), 4722. https://doi.org/10.3390/ijms20194722