The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice
Abstract
:1. Introduction
2. Results
2.1. The Effects of Low-Temperature Stress on Seed Germination, Growth, and Survival of Three Rice Cultivars Seedlings
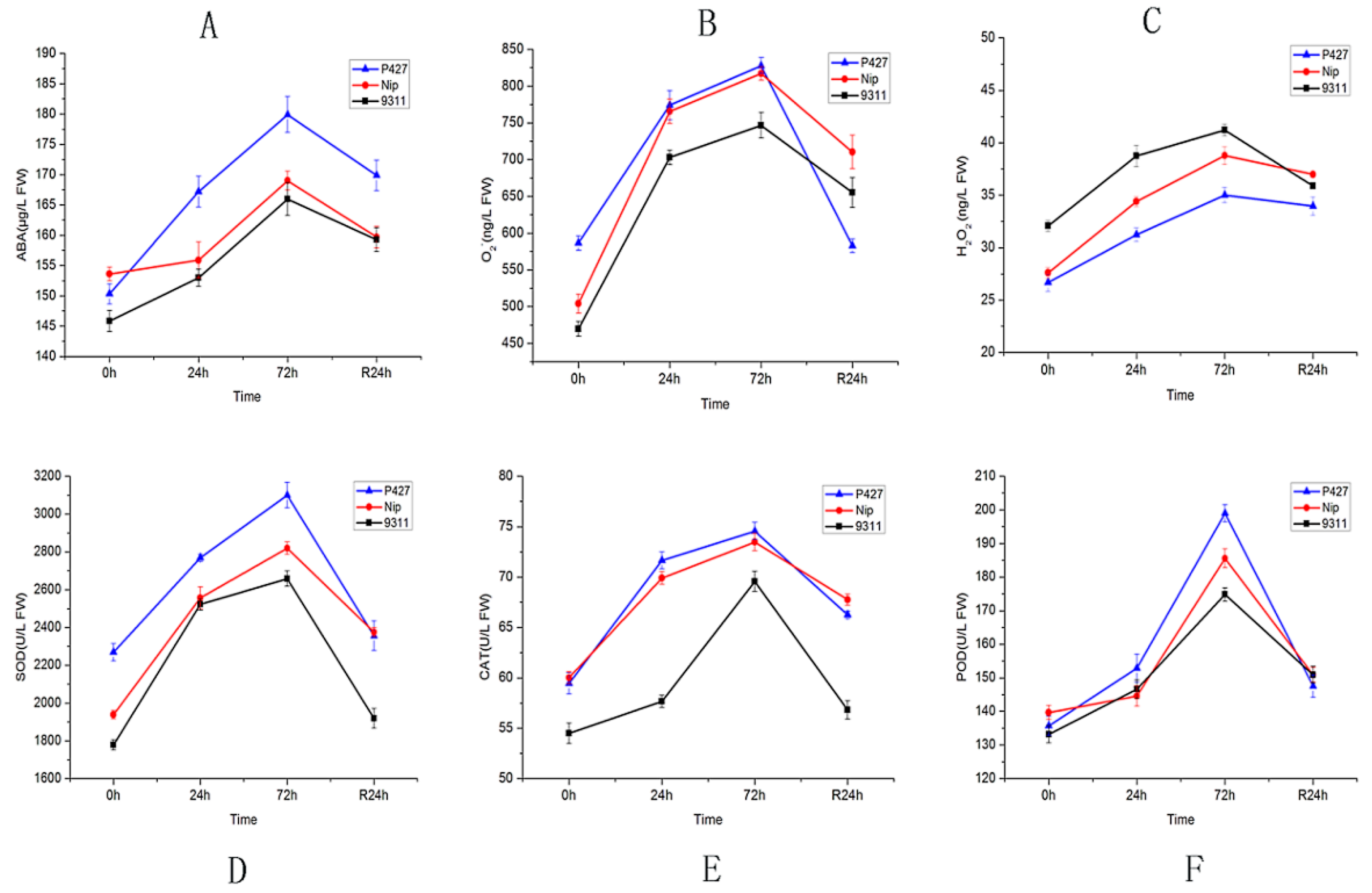
2.2. Physiological Response of Seedlings to Low Temperature
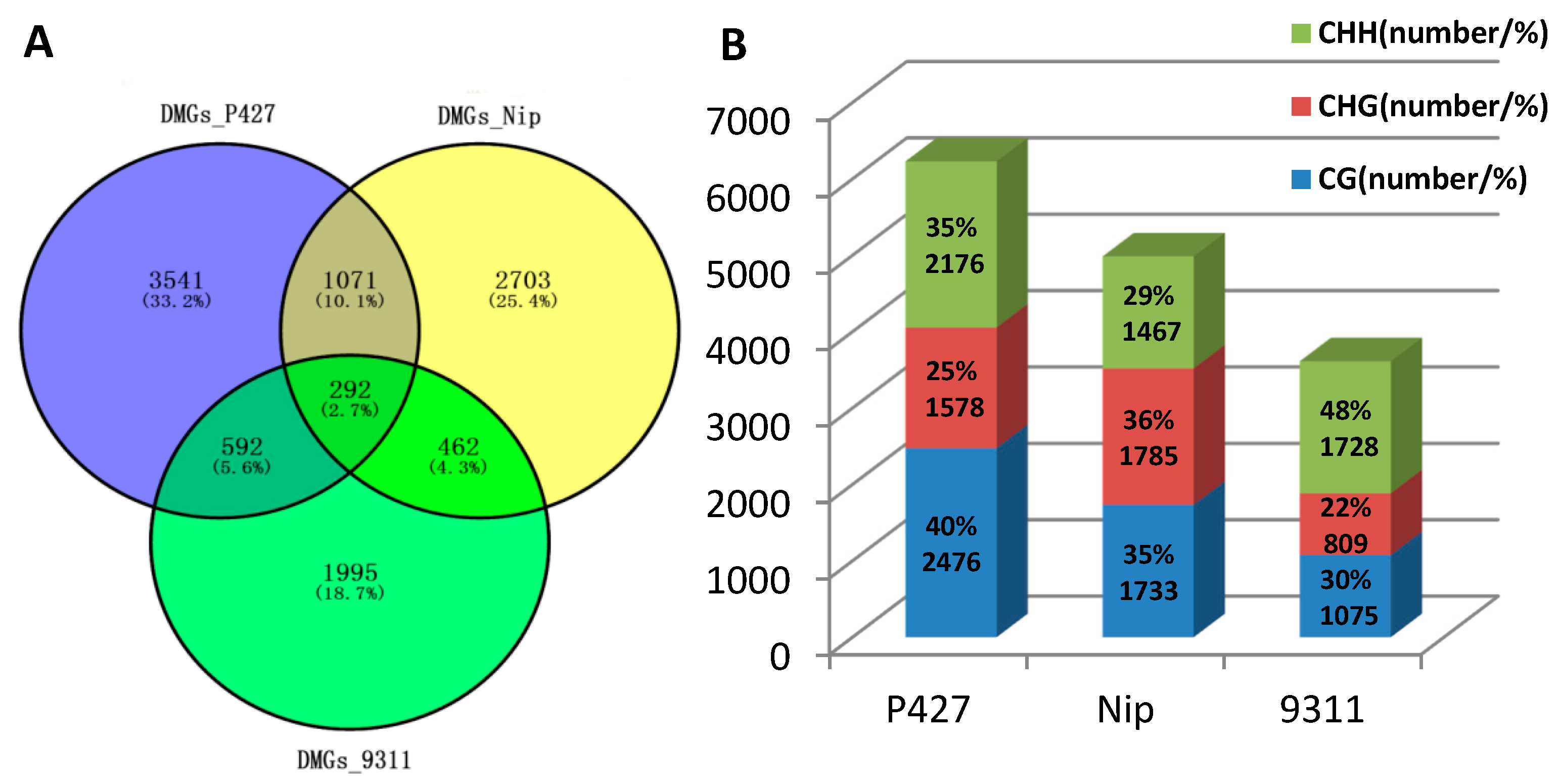
2.3. Comparison of DNA Methylation Levels and Loci
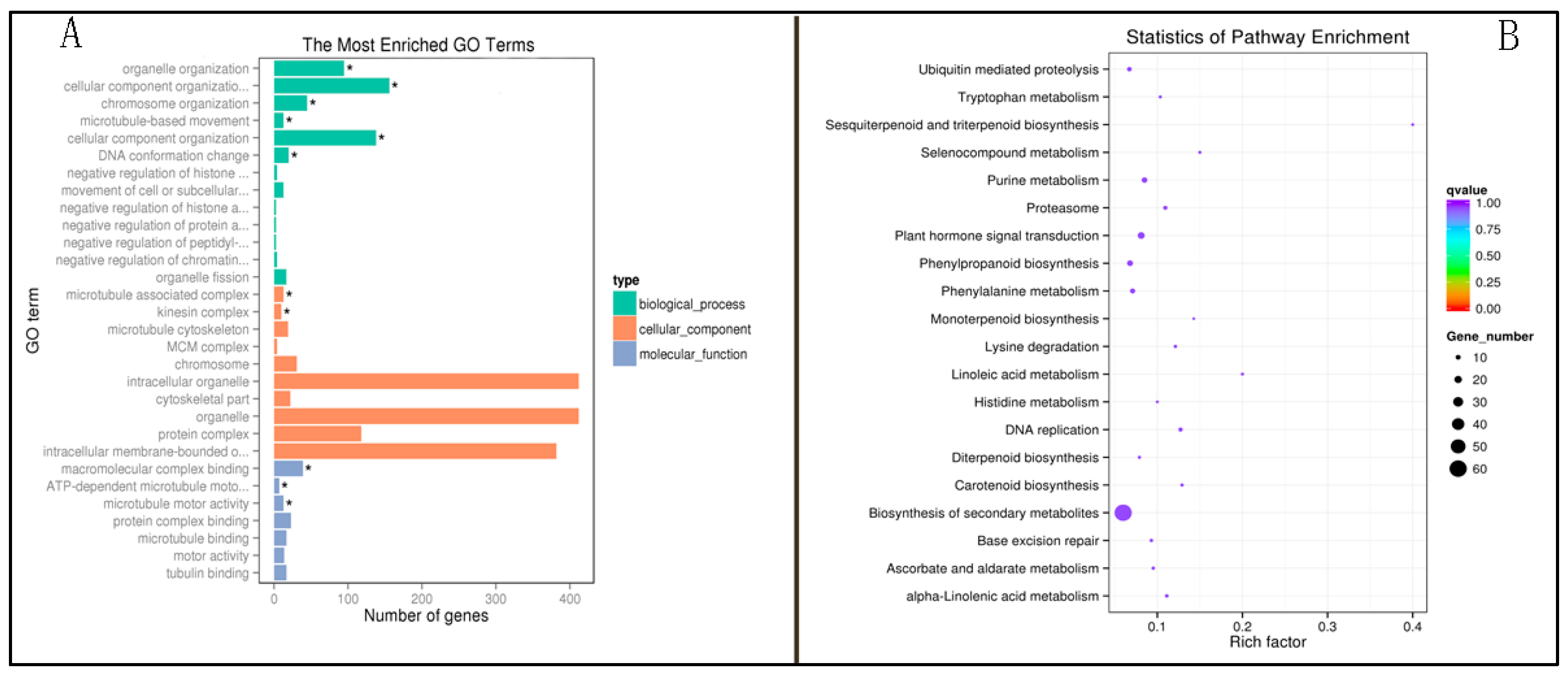
2.4. Functional Enrichment Analysis of the Methylation Differential Gene of P427
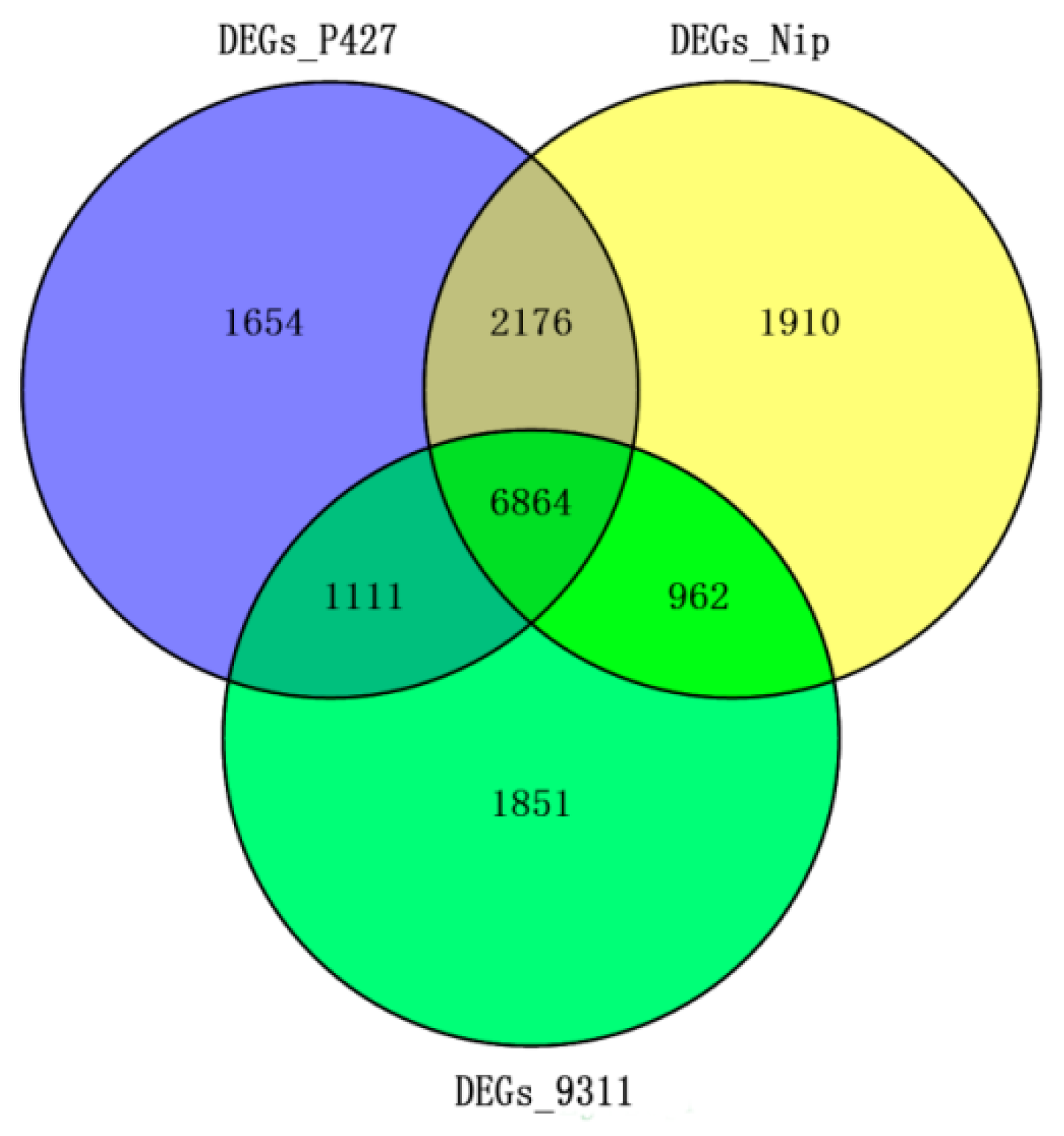
2.5. Comparison of mRNA Expression Differences Under Low-Temperature Stress
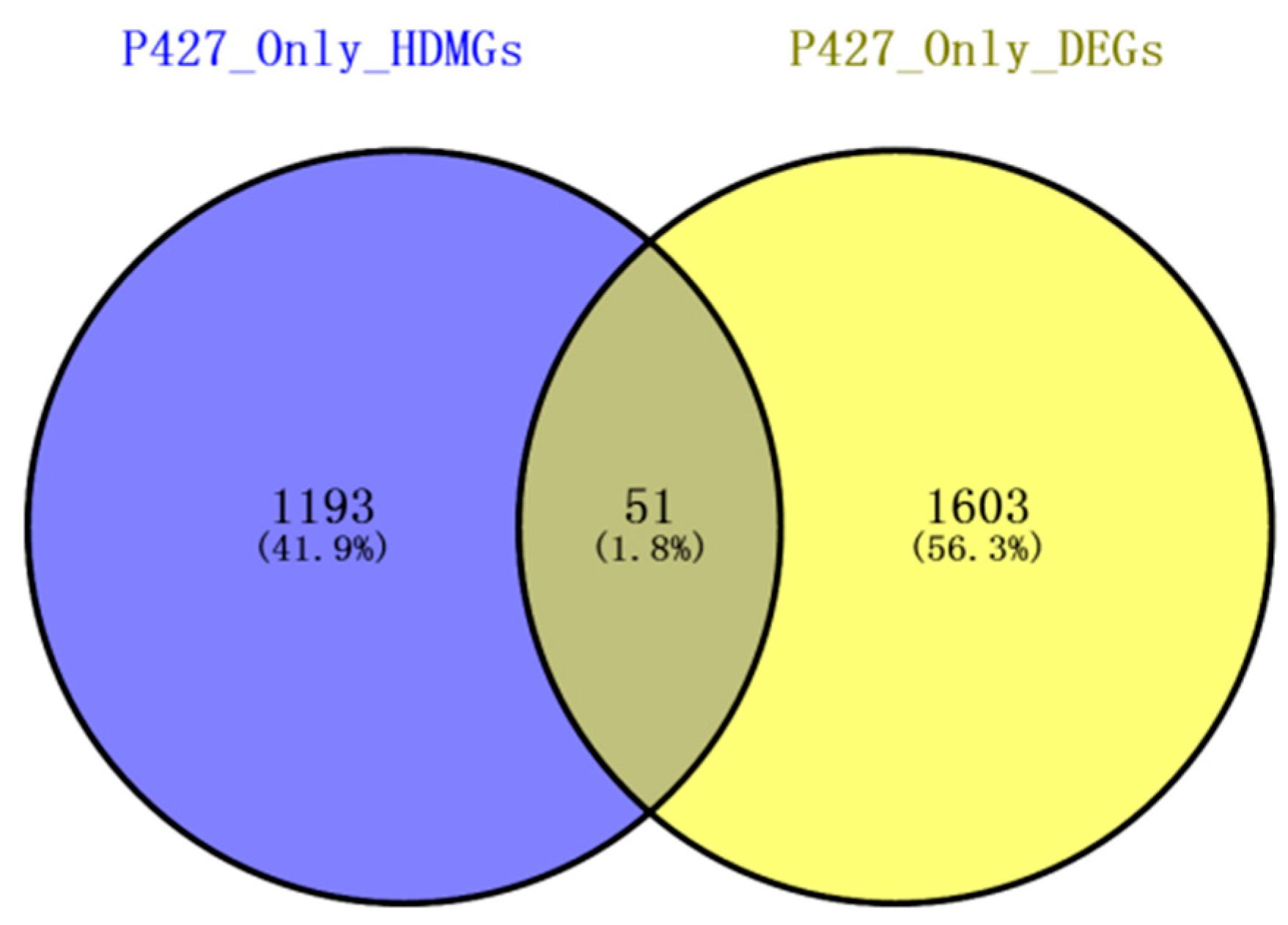
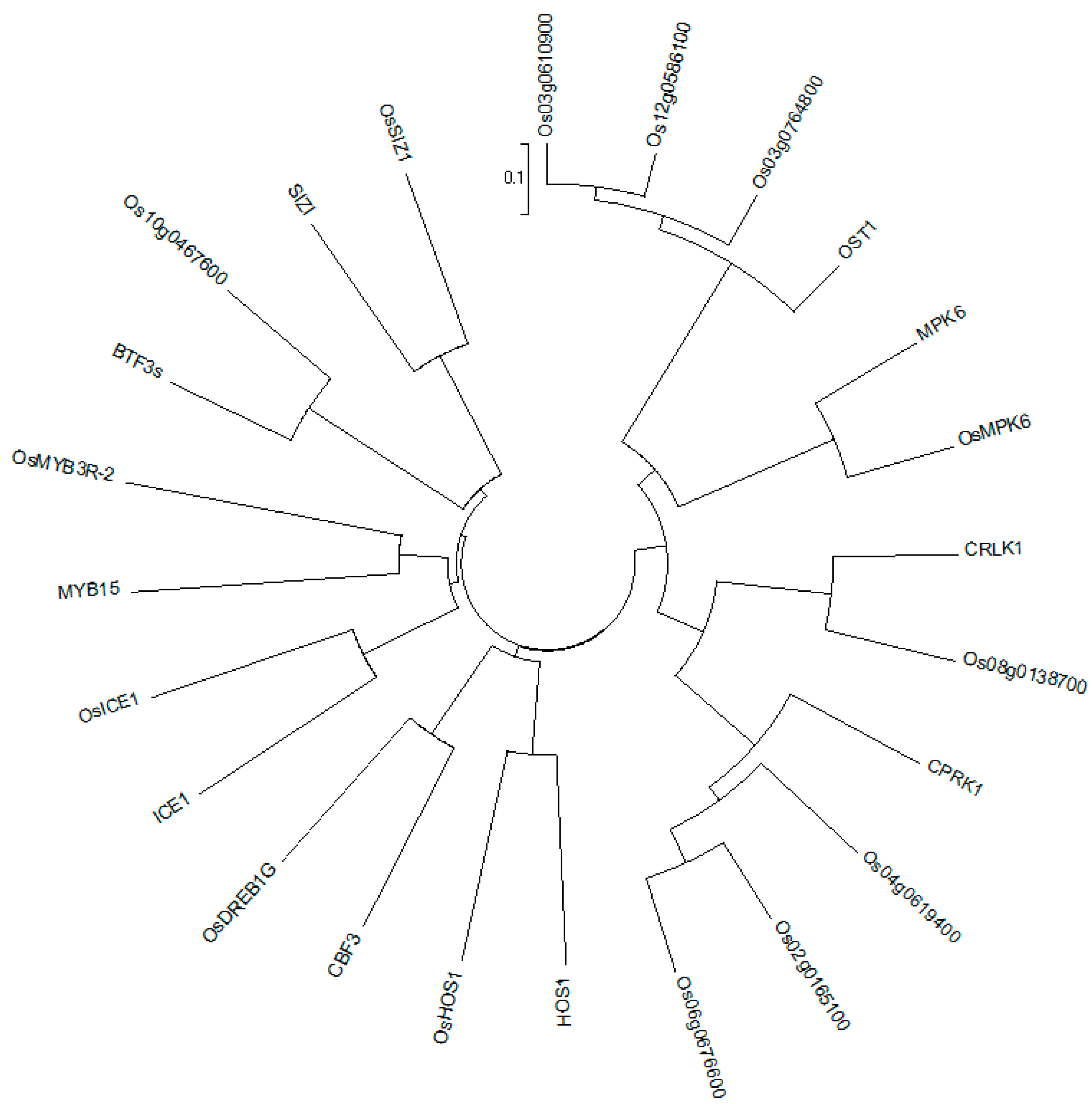
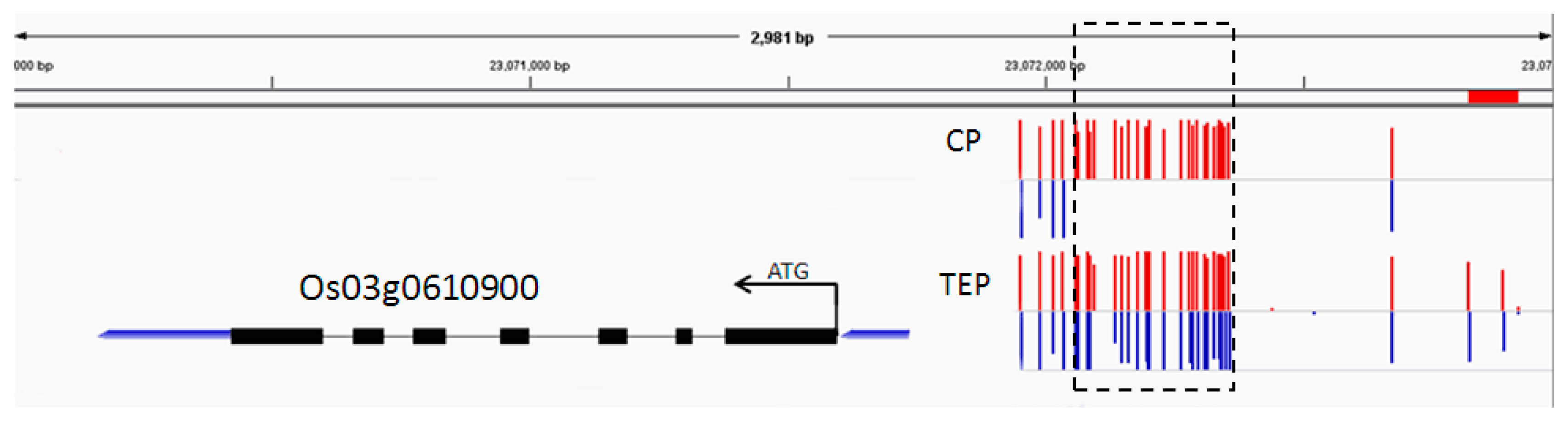
2.6. Combined Analysis of The Methylation and Expression Profiles of Unique Genes in P427
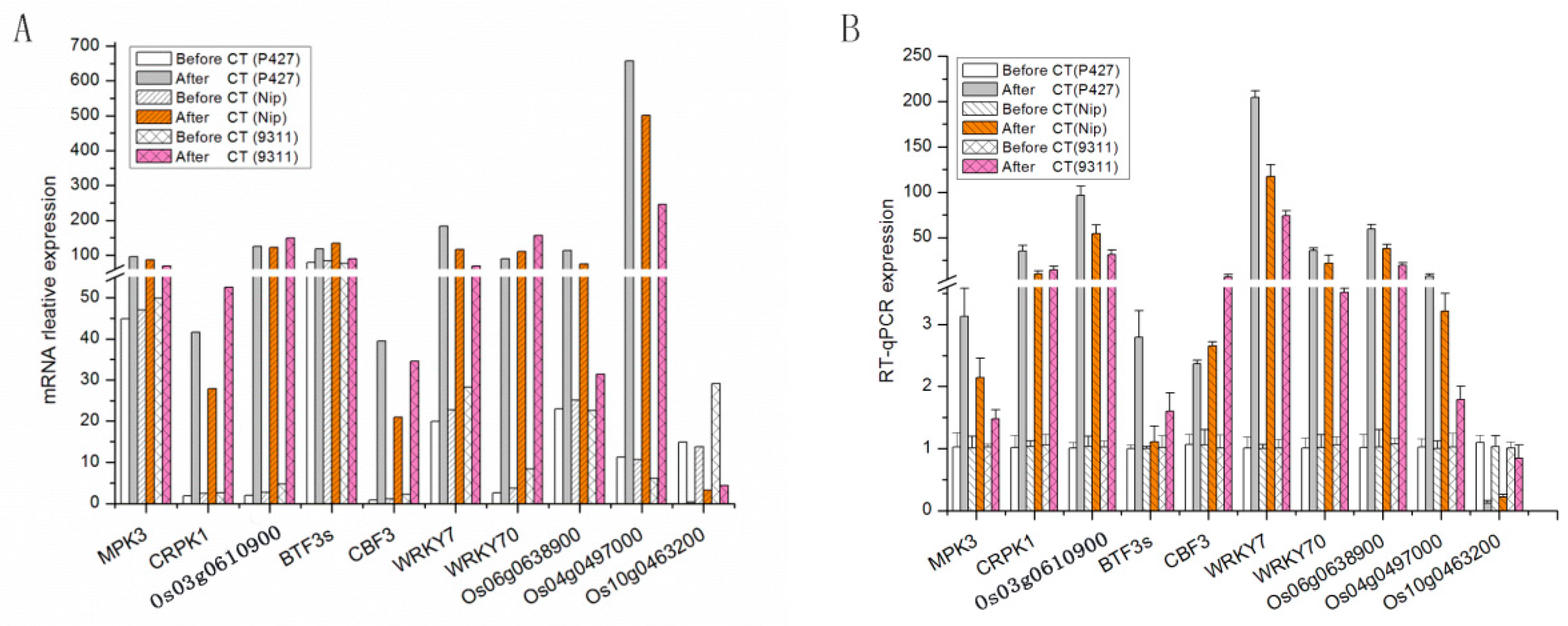
2.7. Validation of Gene Expression Profiles by qRT-PCR
3. Discussion
3.1. The ICE-CBF-COR Conduction Pathway Plays a Leading Role in The Cold Tolerance of Varieties
3.2. Association Analysis between DNA Methylation and Gene Expression
3.3. Excavating New Gene-Rich Cold-Tolerant Regulatory Pathways
4. Materials and Methods
4.1. Low-Temperature Resistance Screening of P427
4.2. Planting and Low-Temperature Treatment
4.3. Chlorophyll Content Determination
4.4. Hormone and Enzyme Content Determination
4.5. RNA-Seq Assay
4.6. MeDIP (Methylated DNA Immunoprecipitation Sequencing) Analysis
4.7. Real-Time Quantitative PCR Validation of The DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chinnusamy, V.; Zhu, J. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant Cold Acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 Have a Different Function than CBF2 in Cold Acclimation and Define Different Gene Classes in the CBF Regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis Cold-Responsive Transcriptome and Its Regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef]
- Xiang, D.J.; Hu, X.Y.; Zhang, Y.; Yin, K.D. Over-pression of ICE1 gene in transgenic rice improve cold tolerance. Rice Sci. 2008, 15, 173–178. [Google Scholar] [CrossRef]
- Xie, H.J.; Li, H.; Liu, D.; Dai, W.M.; He, J.Y.; Lin, S.; Duan, H.; Liu, L.L.; Chen, S.G.; Song, X.L.; et al. ICE1 demethylation drives the range expression of a plant invader through cold tolerance divergence. Mol. Ecol. 2015, 24, 835–850. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 Kinase Modulates Freezing Tolerance by Enhancing ICE1 Stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3- and MPK6-Mediated ICE1 Phosphorylation Negatively Regulates ICE1 Stability and Freezing Tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.-C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP Kinase Cascades Regulate the Cold Response by Modulating ICE1 Protein Stability. Dev. Cell 2017, 43, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Li, F.; Liu, H.; Yang, W.; Chong, K.; Xu, Y. OsMAPK3 Phosphorylates OsbHLH002/OsICE1 and Inhibits Its Ubiquitination to Activate, OsTPP1, and Enhances Rice Chilling Tolerance. Dev. Cell 2017, 43, 731–743.e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jia, Y.; Ding, Y.; Shi, Y.; Li, Z.; Guo, Y.; Gong, Z.; Yang, S. Plasma Membrane CRPK1-Mediated Phosphorylation of 14-3-3 Proteins Induces Their Nuclear Import to Fine-Tune CBF Signaling during Cold Response. Mol. Cell 2017, 66, 117–128.e5. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Zhao, X.; Wang, W.; Pan, Y.; Huang, L.; Liu, X.; Zong, Y.; Zhu, L.; Yang, D.; Fu, B. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS ONE 2012, 7, e43274. [Google Scholar] [CrossRef]
- Miguel, C.; Marum, L. An epigenetic view of plant cells cultured in vitro: Somaclonal variation and beyond. J. Exp. Bot. 2011, 62, 3713–3725. [Google Scholar] [CrossRef]
- De-La-Peña, C.; Nic-Can, G.; Ojeda, G.; Herrera-Herrera, J.L.; López-Torres, A.; Wrobel, K.; Robert-Díaz, M.L. KNOX1 is expressed and epigenetically regulated during in vitro conditions in Agave spp. BMC Plant Biol. 2012, 12, 203. [Google Scholar] [CrossRef]
- Yang, X.; Wang, L.; Yuan, D.; Lindsey, K.; Zhang, X. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 2013, 64, 1521–1536. [Google Scholar] [CrossRef]
- Sahu, P.P.; Pandey, G.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013, 32, 1151–1159. [Google Scholar] [CrossRef]
- Karan, R.; Deleon, T.; Biradar, H.; Subudhi, P.K. Salt Stress Induced Variation in DNA Methylation Pattern and Its Influence on Gene Expression in Contrasting Rice Genotypes. PLoS ONE 2012, 7, e40203. [Google Scholar] [CrossRef]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollander, J.; Meins Jr, F.; Kovalchuk, I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Steward, N.; Ito, M.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002, 277, 37741–37746. [Google Scholar] [CrossRef] [PubMed]
- Yanez, B.E.; Wrobel, K.; Lopez, T.A.; Gutiérrez, C.F.; Wrobel, K. Application of reversed-phase high-performance liquid chromatography with fluorimetric detection for simultaneous assessment of global DNA and total RNA methylation in Lepidium sativum: Effect of plant exposure to Cd(II) and Se(IV). Anal. Bioanal. Chem. 2013, 405, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.N.; Kitamura, K.; Mikami, T.; Kishima, Y. Temperature shift coordinately changes the activity and the methylation state of transposon Tam3 in Antirrhinum majus. Plant Physiol. 2003, 132, 1207–1216. [Google Scholar] [CrossRef]
- Tan, M.P. Analysis of DNA methylation of maize in response to osmotic and salt stress based on methylation-sensitive amplified polymorphism. Plant Physiol. Biochem. 2010, 48, 21–26. [Google Scholar] [CrossRef]
- Szyf, M. DNA Methylation, Behavior and Early Life Adversity. J. Genet. Genom. 2013, 40, 331–338. [Google Scholar] [CrossRef]
- Yang, M.N.; Yang, G.L.; Guo, T.; Liu, Y.Z.; Zhang, J.G.; Chen, Z.Q.; Wang, H. DNA methylation under stresses and its prospects in plant drought-resistant breeding. Chin. Agric. Sci. Bull. 2013, 29, 6–11. [Google Scholar]
- Lister, R.; O’malley, R.C.; Tonti-Fillippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef]
- Yan, H.; Kikuchi, S.; Neumann, P.; Zhang, W.; Wu, Y.; Chen, F.; Jiang, J. Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 2010, 63, 353–365. [Google Scholar] [CrossRef]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef]
- Ding, Y.; Jia, Y.; Shi, Y.; Zhang, X.; Song, C.; Gong, Z.; Yang, S. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 2018, 37, e98228. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.L.; Lv, J.; Shi, Y.T.; Gao, J.P.; Hua, J.; Song, C.P.; Gong, Z.Z.; Yang, S. EGR2 phosphatase regulates OST1 kinase activity and freezing tolerance in Arabidopsis. EMBO J. 2019, 38, e99819. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.-L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide highresolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; He, K.; Ma, Y.; Su, N.; He, H.; Stolc, V.; Tongprasit, W.; Jin, W.; Jiang, J.; et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 2008, 20, 259–276. [Google Scholar] [CrossRef]
- KJunya, M.; Kazuo, S.; Kazuko, Y.S. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar]
- Licausi, F.; Ohmetakagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Liu, J.Y.; Dong, C.J. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 2010, 10, 47. [Google Scholar]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fu, J.H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.A.; Farré, E.M.; Thomashow, M.F. Circadian Clock-Associated 1 and Late Elongated Hypocotyl regulate expression of the C-Repeat Binding Factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB Gene, OsMYB3R-2, Increases Tolerance to Freezing, Drought, and Salt Stress in Transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Su, C.F.; Wang, Y.C.; Hsieh, T.-H.; Lu, C.-A.; Tseng, T.-H.; Yu, S.-M. A Novel MYBS3-Dependent Pathway Confers Cold Tolerance in Rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Zhao, M.-L.; Wang, J.-N.; Shan, W.; Fan, J.G.; Kuang, J.-F.; Wu, K.-Q.; Li, X.-P.; Chen, W.-X.; He, F.-Y.; Chen, J.-Y.; et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2012, 36, 30–51. [Google Scholar] [CrossRef]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2010, 72, 567–568. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Hénanff, G.L.; Profizi, C.; Courteaux, B.; Rabenoelina, F.; Gérard, C.; Clément, C.; Baillieul, F.; Cordelier, S.; Dhondt-Cordelier, S. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013, 64, 4877–4893. [Google Scholar] [CrossRef] [Green Version]
- Ciftciyilmaz, S.; Mittler, R. The zinc finger network of plants. Cell. Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sun, S.; Xu, D.; Lan, H.; Sun, H.; Wang, Z.; Bao, Y.; Wang, J.; Tang, H.; Zhang, H. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol. 2012, 80, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ouyang, B.; Zhang, J.; Wang, T.; Li, H.; Zhang, Y.; Yu, C.; Ye, Z. Differential Modulation of Photosynthesis, Signaling, and Transcriptional Regulation between Tolerant and Sensitive Tomato Genotypes under Cold Stress. PLoS ONE 2012, 7, e50785. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.-S.; Chen, S.-Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Shi, Y.T.; Yang, S.H. CBF: A Key Factor Balancing Plant Cold Stress Responses and Growth. Chin. Bull. Bot. 2017, 52, 689–698. [Google Scholar]
- Chawade, A.; Lindlöf, A.; Olsson, B.; Olsson, O. Global Expression Profiling of Low Temperature Induced Genes in the Chilling Tolerant Japonica Rice Jumli Marshi. PLoS ONE 2013, 8, e81729. [Google Scholar] [CrossRef]
- Zhang, L.; Gu, L.; Ringler, P.; Smith, S.; Rushton, P.J.; Shen, Q.J. Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 2015, 236, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Barrero-Gil, J.V.; Salinas, J. Post-translational regulation of cold acclimation response. Plant Sci. 2013, 205, 48–54. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Wu, T.; Li, S.; He, Q.; Yang, Z.; Zhang, W.; Gan, Y.; Sun, P.; Xiang, G.; Zhang, H.; et al. The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice. Int. J. Mol. Sci. 2019, 20, 5089. https://doi.org/10.3390/ijms20205089
Guo H, Wu T, Li S, He Q, Yang Z, Zhang W, Gan Y, Sun P, Xiang G, Zhang H, et al. The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice. International Journal of Molecular Sciences. 2019; 20(20):5089. https://doi.org/10.3390/ijms20205089
Chicago/Turabian StyleGuo, Hui, Tingkai Wu, Shuxing Li, Qiang He, Zhanlie Yang, Wuhan Zhang, Yu Gan, Pingyong Sun, Guanlun Xiang, Hongyu Zhang, and et al. 2019. "The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice" International Journal of Molecular Sciences 20, no. 20: 5089. https://doi.org/10.3390/ijms20205089



