Epigenetics, Stem Cells, and Autophagy: Exploring a Path Involving miRNA
Abstract
:1. Introduction
2. Results
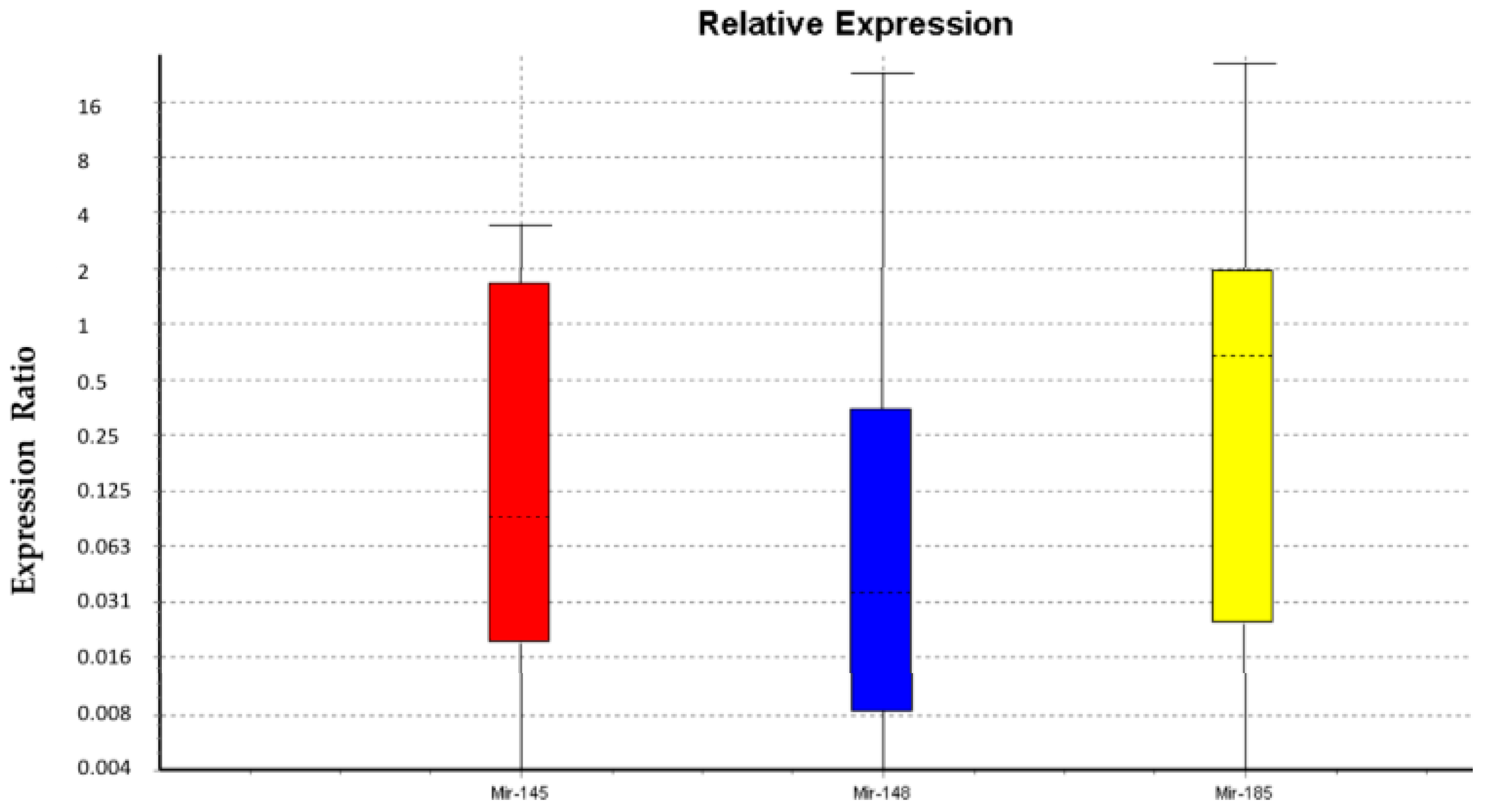
2.1. miRNA Expression in WJ-MSCs and Gender Differences
2.2. Adipogenic and Osteogenic Differentiation is Similar in Males and Females WJ-MSCs
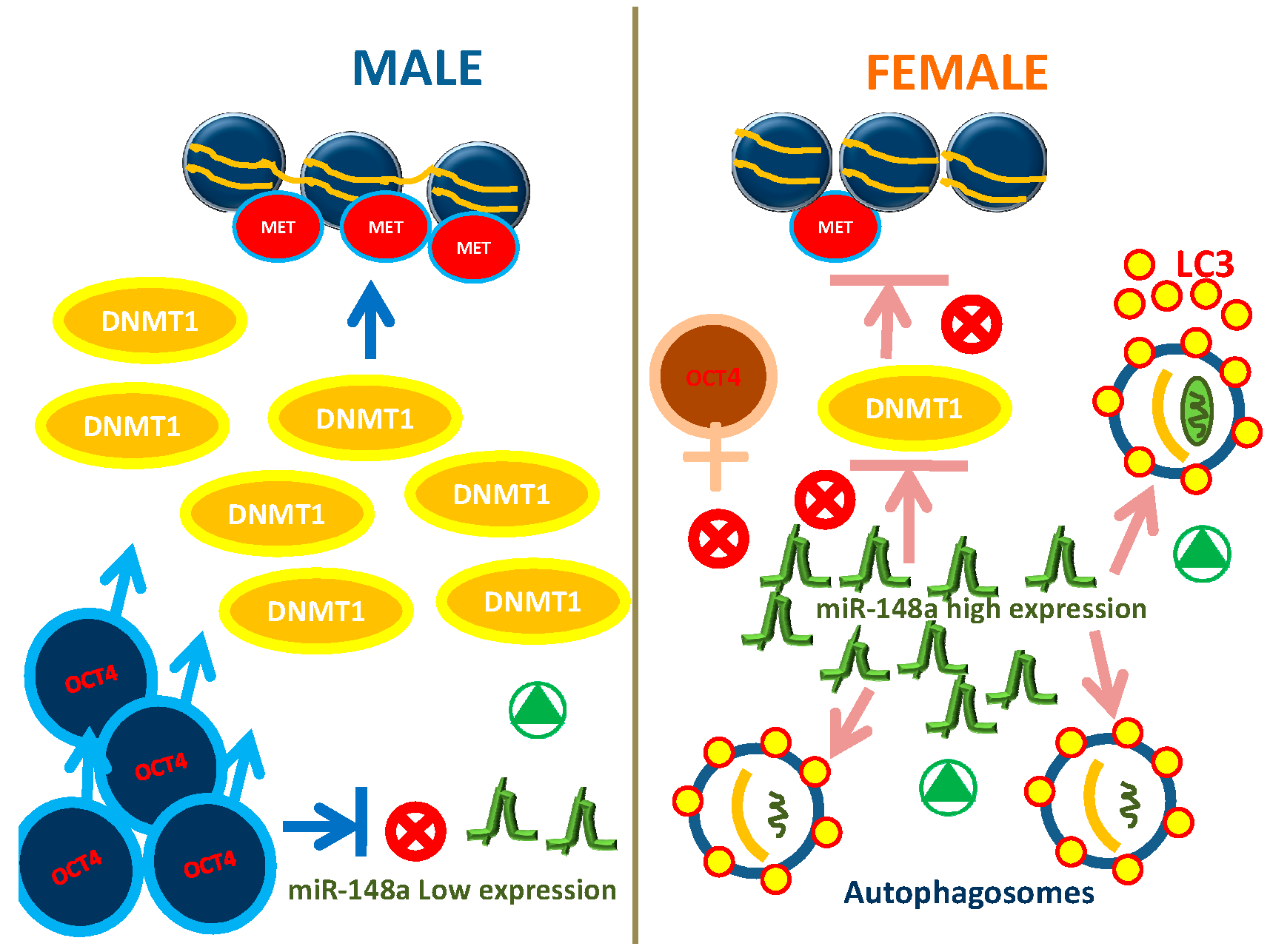
2.3. Autophagy Exhibit Gender Differences in WJ-MSCs
3. Discussion
4. Materials and Methods
4.1. WJ-MSCs Isolation and Culture
4.1.1. RNA Extraction and Quantitative Polymerase Chain Reaction
4.1.2. List of Software for the Identification of miRNAs
4.1.3. Quantitative PCR Analysis
4.1.4. Statistical Analysis and Real-Time PCR Data Analysis
4.2. Analysis of Cellular Mechanisms in WJ-MSCs from Males and Females
4.2.1. Osteogenic Differentiation: Alizarin Red Assay
4.2.2. Adipogenic Differentiation: Red Oil Assay
4.3. Autophagy Detection: Western Blotting
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | linear dichroism |
References
- Balzano, F.; Cruciani, S.; Basoli, V.; Santaniello, S.; Facchin, F.; Ventura, C.; Maioli, M. MiR200 and miR302: Two Big Families Influencing Stem Cell Behavior. Molecules 2018, 23, 282. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef]
- Rinaldi, S.; Maioli, M.; Pigliaru, G.; Castagna, A.; Santaniello, S.; Basoli, V.; Fontani, V.; Ventura, C. Stem cell senescence. Effects of REAC technology on telomerase-independent and telomerase-dependent pathways. Sci. Rep. 2014, 4, 6373. [Google Scholar] [CrossRef] [Green Version]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Gualini, S.; Cavallini, C.; Fontani, V.; Ventura, C. Radio Electric Conveyed Fields Directly Reprogram Human Dermal Skin Fibroblasts toward Cardiac, Neuronal, and Skeletal Muscle-Like Lineages. Cell Transplant. 2013, 22, 1227–1235. [Google Scholar] [CrossRef] [Green Version]
- Balzano, F.; Bellu, E.; Basoli, V.; Dei Giudici, S.; Santaniello, S.; Cruciani, S.; Facchin, F.; Oggiano, A.; Capobianco, G.; Dessole, F.; et al. Lessons from human umbilical cord: Gender differences in stem cells from Wharton’s jelly. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 234, 143–148. [Google Scholar] [CrossRef]
- Schaefer, M.; Lyko, F. DNA methylation with a sting: An active DNA methylation system in the honeybee. BioEssays 2007, 29, 208–211. [Google Scholar] [CrossRef]
- Imamura, M.; Miura, K.; Iwabuchi, K.; Ichisaka, T.; Nakagawa, M.; Lee, J.; Kanatsu-Shinohara, M.; Shinohara, T.; Yamanaka, S. Transcriptional repression and DNA hypermethylationof a small set of ES cell marker genes in male germline stem cells. BMC Dev. Biol. 2006, 6, 34. [Google Scholar] [CrossRef]
- Saini, S.K.; Mangalhara, K.C.; Prakasam, G.; Bamezai, R.N.K. DNA Methyltransferase1 (DNMT1) Isoform3 methylates mitochondrial genome and modulates its biology. Sci. Rep. 2017, 7, 1525. [Google Scholar] [CrossRef] [PubMed]
- Yeom, Y.I.; Fuhrmann, G.; Ovitt, C.E.; Brehm, A.; Ohbo, K.; Gross, M.; Hübner, K.; Schöler, H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 1996, 122, 881–894. [Google Scholar] [PubMed]
- Tsai, C.-C.; Su, P.F.; Huang, Y.-F.; Yew, T.L.; Hung, S.C. Oct4 and Nanog Directly Regulate Dnmt1 to Maintain Self-Renewal and Undifferentiated State in Mesenchymal Stem Cells. Mol. Cell 2012. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Qu, H.; Qi, D.; Xu, W.; Liu, S.; Jin, X.; Song, P.; Guo, Y.; Jia, Y.; Wang, X.; et al. OCT4 maintains self-renewal and reverses senescence in human hair follicle mesenchymal stem cells through the downregulation of p21 by DNA methyltransferases. Stem Cell Res. Ther. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.E.; Hu, F.W.; Yu, C.H.; Tsai, L.L.; Lee, T.H.; Chou, M.Y.; Yu, C.C. Concurrent Expression of Oct4 and Nanog Maintains Mesenchymal Stem-Like Property of Human Dental Pulp Cells. Int. J. Mol. Sci. 2014, 15, 18623–18639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Delitala, A.; Bianchi, F.; Tremolada, C.; Fontani, V.; Ventura, C. Radioelectric Asymmetric Conveyed Fields and Human Adipose-Derived Stem Cells Obtained with a Nonenzymatic Method and Device: A Novel Approach to Multipotency. Cell Transplant. 2014, 23, 1489–1500. [Google Scholar] [CrossRef]
- Maioli, M.; Rinaldi, S.; Migheli, R.; Pigliaru, G.F.; Rocchitta, G.; Santaniello, S.; Basoli, V.; Castagna, A.; Fontani, V.; Ventura, C.; et al. Neurological morpho functional differentiation induced by REAC technology in PC12. A neuro protective model for Parkinson’s disease. Sci. Rep. 2015, 5, 10439. [Google Scholar] [CrossRef]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Gualini, S.; Fontani, V.; Ventura, C. Radiofrequency Energy Loop Primes Cardiac, Neuronal, and Skeletal Muscle Differentiation in Mouse Embryonic Stem Cells: A New Tool for Improving Tissue Regeneration. Cell Transplant. 2012, 21, 1225–1233. [Google Scholar] [CrossRef] [Green Version]
- Maioli, M.; Rinaldi, S.; Pigliaru, G.; Santaniello, S.; Basoli, V.; Castagna, A.; Fontani, V.; Ventura, C. REAC technology and hyaluron synthase 2, an interesting network to slow down stem cell senescence. Sci. Rep. 2016, 6, 28682. [Google Scholar] [CrossRef] [Green Version]
- Santaniello, S.; Cruciani, S.; Basoli, V.; Balzano, F.; Bellu, E.; Garroni, G.; Ginesu, G.C.; Cossu, M.L.; Facchin, F.; Delitala, A.P.; et al. Melatonin and Vitamin D Orchestrate Adipose Derived Stem Cell Fate by Modulating Epigenetic Regulatory. Genes Int. J. Med. Sci. 2018, 15, 1631–1639. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Kuma, A.; Hatano, M.; Matsui, M.; Yamamoto, A.; Nakaya, H.; Yoshimori, T.; Ohsumi, Y.; Tokuhisa, T.; Mizushima, N. The role of autophagy during the early neonatal starvation period. Nature 2004, 432, 1032–1036. [Google Scholar] [CrossRef]
- Hong, L.; Sun, G.; Peng, L.; Tu, Y.; Wan, Z.; Xiong, H.; Li, Y.; Xiao, W. The interaction between miR-148a and DNMT1 suppresses cell migration and invasion by reactivating tumor suppressor genes in pancreatic cancer. Oncol. Rep. 2018, 40, 2916–2925. [Google Scholar] [CrossRef]
- Lopez-Bertoni, H.; La, B.; Li, A.; Caplan, M.; Guerrero-Cázares, H.; Eberhart, C.G.; Quiñones-Hinojosa, A.; Glas, M.; Scheffler, B.; Laterra, J.; et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 2015, 34, 3994–4004. [Google Scholar] [CrossRef]
- Wang, P.; Li, X.; Cao, L.; Huang, S.; Li, H.; Zhang, Y.; Yang, T.; Jiang, J.; Shi, D. MicroRNA-148a overexpression improves the early development of porcine somatic cell nuclear transfer embryos. PLoS ONE 2017, 12, e0180535. [Google Scholar] [CrossRef]
- Liu, X.-Y.; He, Y.-J.; Yang, Q.-H.; Huang, W.; Liu, Z.-H.; Ye, G.-R.; Tang, S.-H.; Shu, J.-C. Induction of autophagy and apoptosis by miR-148a through the sonic hedgehog signaling pathway in hepatic stellate cells. Am. J. Cancer Res. 2015, 9, 2569–2589. [Google Scholar]
- Menghini, R.; Casagrande, V.; Marino, A.; Marchetti, V.; Cardellini, M.; Stoehr, R.; Rizza, S.; Martelli, E.; Greco, S.; Mauriello, A.; et al. MiR-216a: A link between endothelial dysfunction and autophagy. Cell Death Dis. 2014, 5, e1029. [Google Scholar] [CrossRef]
- Yeh, Y.; Weiab, J.; Thorossianb, S.; Nguyenab, K.; Hoffmanab, C.; del Álamobc, J.C.; Serranoc, R.; Julie, Y.-S.; Kuei-Chun, L.; Chienab, W.S. MiR-145 mediates cell morphology-regulated mesenchymal stem cell differentiation to smooth muscle. Cells Biomater. 2019, 204, 59–69. [Google Scholar] [CrossRef]
- Cui, Q.; Xing, J.; Yu, M.; Wang, Y.; Xu, J.; Gu, Y.; Nan, X.; Ma, W.; Liu, H.; Zhao, H. Mmu-miR-185 depletion promotes osteogenic differentiation and suppresses bone loss in osteoporosis through the Bgn-mediated BMP/Smad pathway. Cell Death Dis. 2019, 103, 172. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Campesi, I.; Straface, E.; Occhioni, S.; Montella, A.; Franconi, F. Protein oxidation seems to be linked to constitutive autophagy: A sex study. Life Sci. 2013, 93, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Addis, R.; Campesi, I.; Fois, M.; Capobianco, G.; Dessole, S.; Fenu, G.; Montella, A.; Cattaneo, M.G.; Vicentini, L.M.; Franconi, F. Human umbilical endothelial cells (HUVECs) have a sex: Characterisation of the phenotype of male and female cells. Biol. Sex Differ. 2014, 51, 18. [Google Scholar] [CrossRef] [PubMed]
- Campes, I.; Occhioni, S.; Capobianco, G.; Fois, M.; Montella, A.; Dessole, S.; Franconi, F. Sex-specific pharmacological modulation of autophagic process in human umbilical artery smooth muscle cells. Pharmacol. Res. 2016, 113, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Basoli, V.; Santaniello, S.; Cruciani, S.; Ginesu, G.C.; Cossu, M.L.; Delitala, A.P.; Serra, P.A.; Ventura, C.; Maioli, M. Melatonin and Vitamin D Interfere with the Adipogenic Fate of Adipose-Derived Stem Cells. Int. J. Mol. Sci. 2017, 18, 981. [Google Scholar] [CrossRef]
- Maioli, M.; Basoli, V.; Santaniello, S.; Cruciani, S.; Delitala, A.P.; Pinna, R.; Milia, E.; Grillari-Voglauer, R.; Fontani, V.; Rinaldi, S.; et al. Osteogenesis from Dental Pulp Derived Stem Cells: A Novel Conditioned Medium Including Melatonin within a Mixture of Hyaluronic, Butyric, and Retinoic Acids. Stem Cells Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Maioli, M.; Contini, G.; Santaniello, S.; Bandiera, P.; Pigliaru, G.; Sanna, R.; Rinaldi, S.; Delitala, A.P.; Montella, A.; Bagella, L.; et al. Amniotic fluid stem cells morph into a cardiovascular lineage: Analysis of a chemically induced cardiac and vascular commitment. Drug Des. Dev. Ther. 2013, 7, 1063–1073. [Google Scholar]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Delitala, A.; Lotti Margotti, M.; Bagella, L.; Fontani, V.; Ventura, C. Anti-senescence efficacy of radio-electric asymmetric conveyer technology. Age 2014, 1, 9–20. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, Y.; Zhang, Y.; Zhang, Y.; Chen, L.; Mo, D. Up-regulated miR-145 Expression Inhibits Porcine Preadipocytes Differentiation by Targeting IRS1. Int. J. Boil. Sci. 2012, 8, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef] [PubMed]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, L.S.; Tastsoglou, S.; Kanellos, L.; Papadimitriou, D.; Kavakiotis, L.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 4, D239–D245. [Google Scholar] [CrossRef]
- Arderiu, G.; Peña, E.; Aledo, R.; Juan-Babot, O.; Crespo, J.; Vilahur, G.; Onate, B.; Moscatiello, F.; Badimon, L. MicroRNA-145 Regulates the Differentiation of Adipose Stem Cells Toward Microvascular Endothelial Cells and Promotes Angiogenesis. Circ. Res. 2019, 125, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Balzano, F.; Deiana, M.; Giudici, S.D.; Oggiano, A.; Baralla, A.; Pasella, S.; Mannu, A.; Pescatori, M.; Porcu, B.; Fanciulli, G.; et al. miRNA Stability in Frozen Plasma Samples. Molecules 2015, 20, 19030–19040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benz, F.; Roderburg, C.; Cardenas, D.V.; Vucur, M.; Gautheron, J.; Koch, A.; Zimmermann, H.; Janssen, J.; Nieuwenhuijsen, L.; Luedde, M.; et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013, 45, e42. [Google Scholar] [CrossRef] [PubMed]
- miRbase. Available online: http://www.mirbase.org/ (accessed on 24 September 2015).
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) forgroup-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, 9. [Google Scholar] [CrossRef]
- Mazzini, L.; Ferrari, D.; Andjus, P.R.; Buzanska, L.; Cantello, R.; De Marchi, F.; Gelati, M.; Giniatullin, R.; Glover, J.C.; Grilli, M.; et al. Advances in stem cell therapy for amyotrophic lateral sclerosis. Expert Opin. Biol. Ther. 2018, 18, 865–881. [Google Scholar] [CrossRef]


| Gene miRNA Software | Databases |
|---|---|
| DNMT1 | hsa-miR-148a-3p miRTarBase, Targetscan, DIANA-TarBase, miRBD |
| DNMT1 | hsa-miR-185 miRanda, Targetscan, PITA |
| OCT4 | hsa-miR-145-5p miRTarBase, DIANA-TarBase |
| Relative Expression Results | |||||||
|---|---|---|---|---|---|---|---|
| Parameter | Value | ||||||
| Iterations | 2000 | ||||||
| Gene | Type | Reaction Efficiency | Expression | Std. Error | 95% C.I. | P(H1) | Result |
| miR-145 | TRG | 1.0 | 0.099 | 0.009-2.083 | 0.001-3.079 | 0.064 | |
| miR-148 | TRG | 1.0 | 0.059 | 0.005-1.508 | 0.001-22.627 | 0.039 | Down |
| miR-185 | TRG | 1.0 | 0.326 | 0.006-7.205 | 0.001-25.813 | 0.419 | |
| US6 | REF | 1.0 | 1.000 | ||||
| Accession ID Number Symbol Sequence |
|---|
| MIMAT0000243 hsa-miR-148a-3p UCAGUGCACUACAGAACUUUGU |
| MIMAT0004611 hsa-miR-185-3p AGGGGCUGGCUUUCCUCUGGUC |
| MIMAT0000437 hsa-miR-145-5p GUCCAGUUUUCCCAGGAAUCCCU |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balzano, F.; Campesi, I.; Cruciani, S.; Garroni, G.; Bellu, E.; Dei Giudici, S.; Angius, A.; Oggiano, A.; Rallo, V.; Capobianco, G.; et al. Epigenetics, Stem Cells, and Autophagy: Exploring a Path Involving miRNA. Int. J. Mol. Sci. 2019, 20, 5091. https://doi.org/10.3390/ijms20205091
Balzano F, Campesi I, Cruciani S, Garroni G, Bellu E, Dei Giudici S, Angius A, Oggiano A, Rallo V, Capobianco G, et al. Epigenetics, Stem Cells, and Autophagy: Exploring a Path Involving miRNA. International Journal of Molecular Sciences. 2019; 20(20):5091. https://doi.org/10.3390/ijms20205091
Chicago/Turabian StyleBalzano, Francesca, Ilaria Campesi, Sara Cruciani, Giuseppe Garroni, Emanuela Bellu, Silvia Dei Giudici, Andrea Angius, Annalisa Oggiano, Vincenzo Rallo, Giampiero Capobianco, and et al. 2019. "Epigenetics, Stem Cells, and Autophagy: Exploring a Path Involving miRNA" International Journal of Molecular Sciences 20, no. 20: 5091. https://doi.org/10.3390/ijms20205091







