Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.)
Abstract
:1. Introduction
2. Results
2.1. Identification and Chromosome Distribution of bZIP Family in Radish
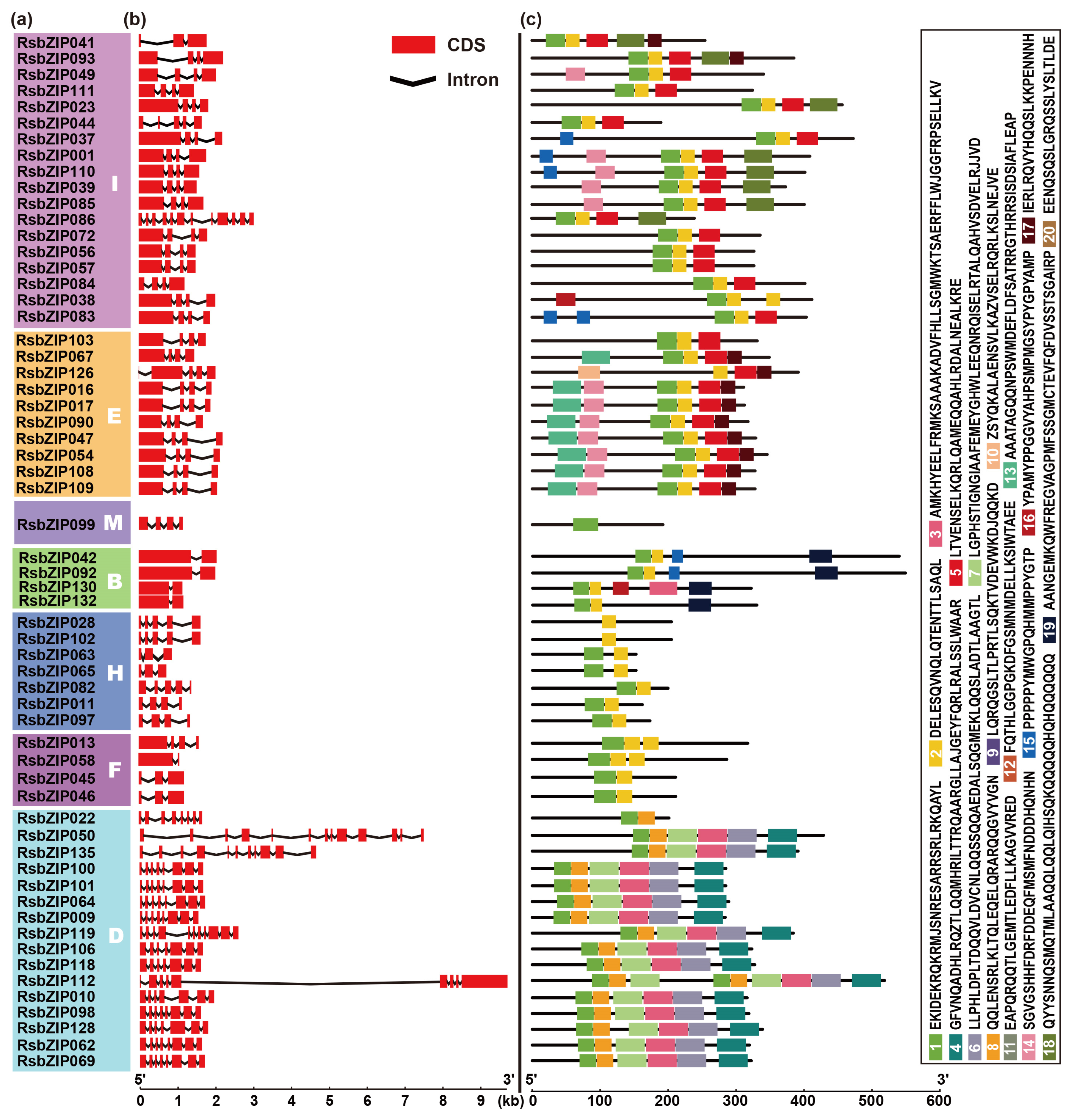
2.2. Gene Structure and Conserved Motif Analysis of RsbZIPs
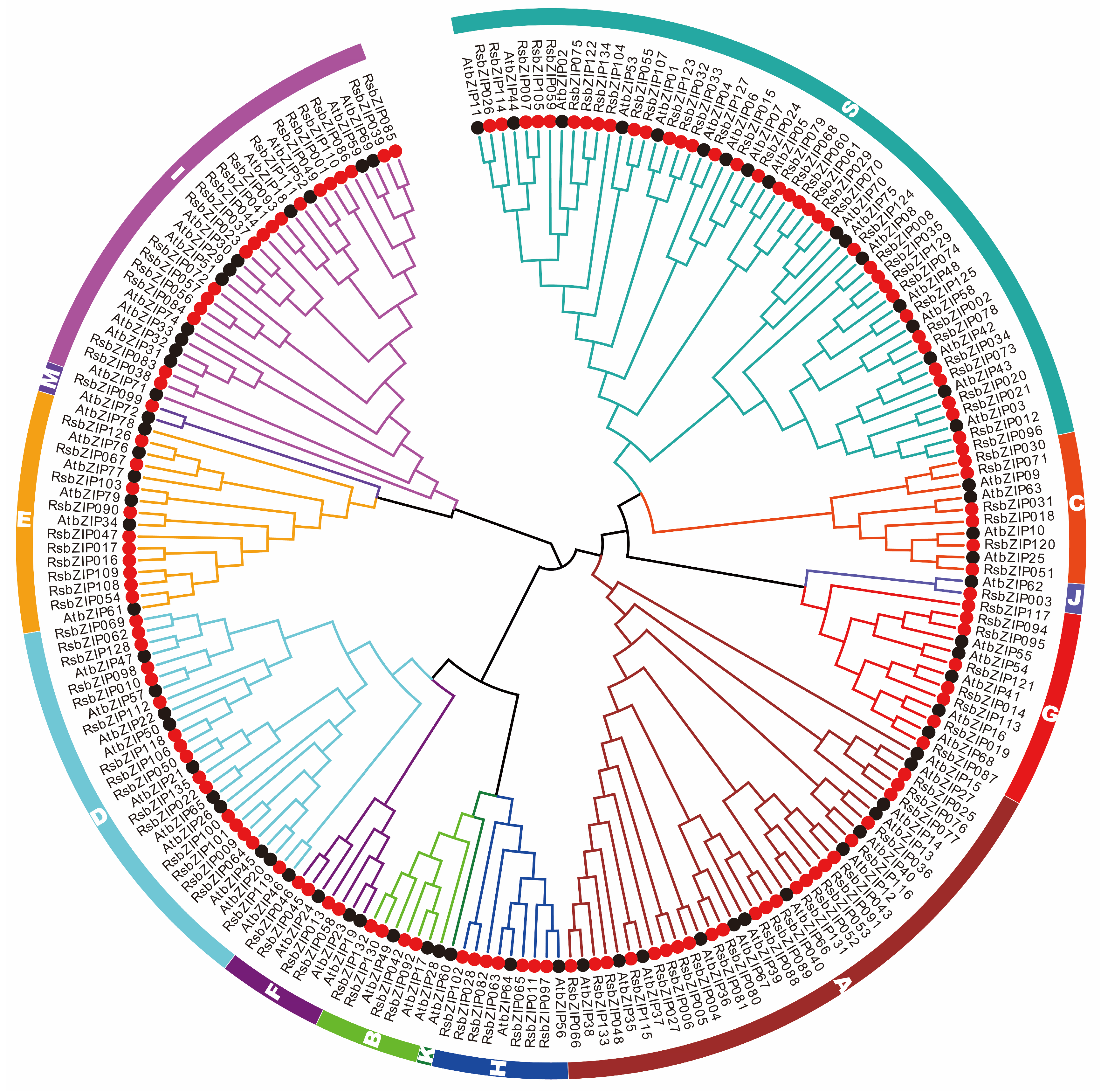
2.3. Phylogenetic and Evolutionary of Radish and Arabidopsis bZIP Genes
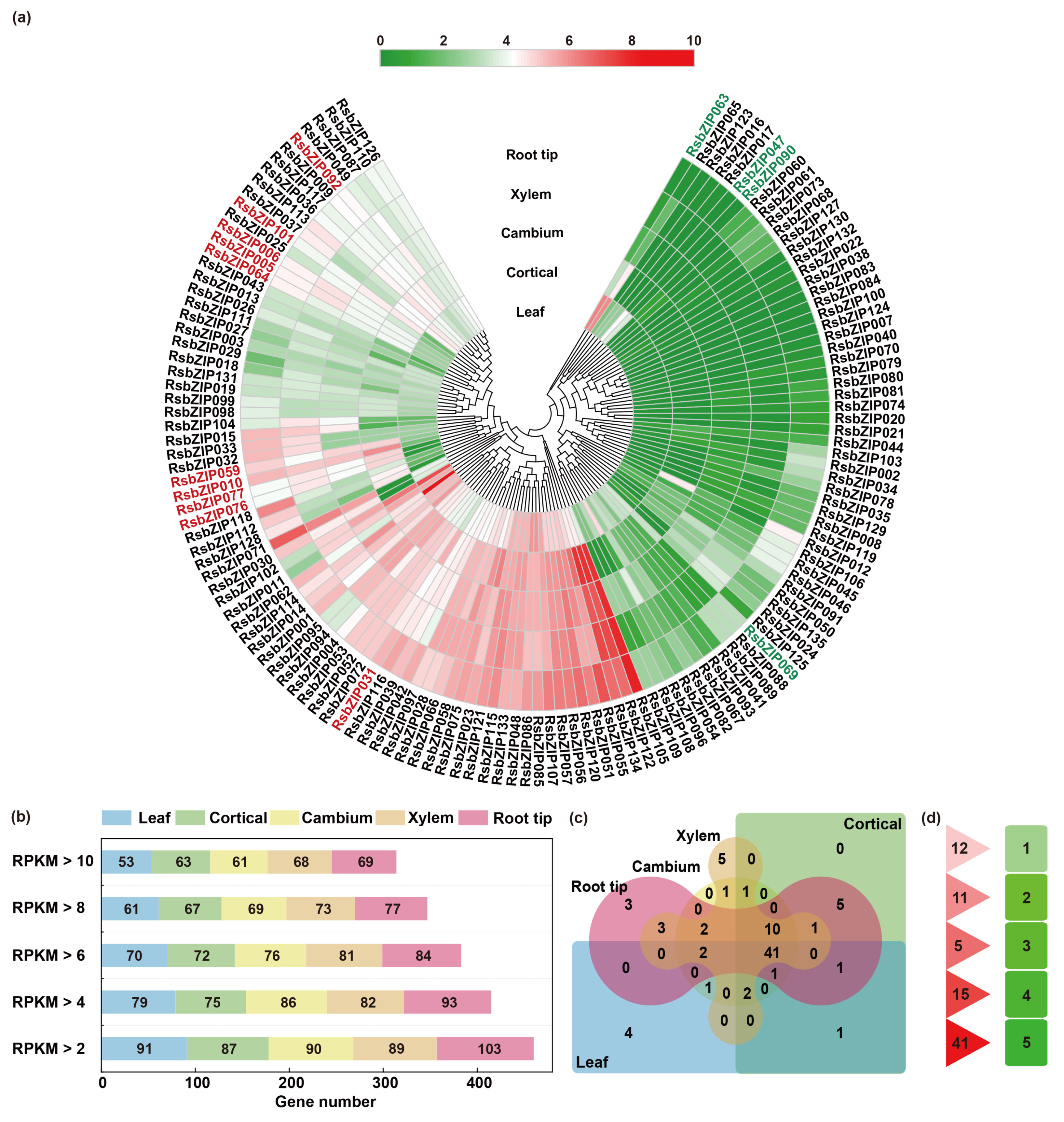
2.4. Expression Profiles of bZIP Genes in Different Tissues of Radish
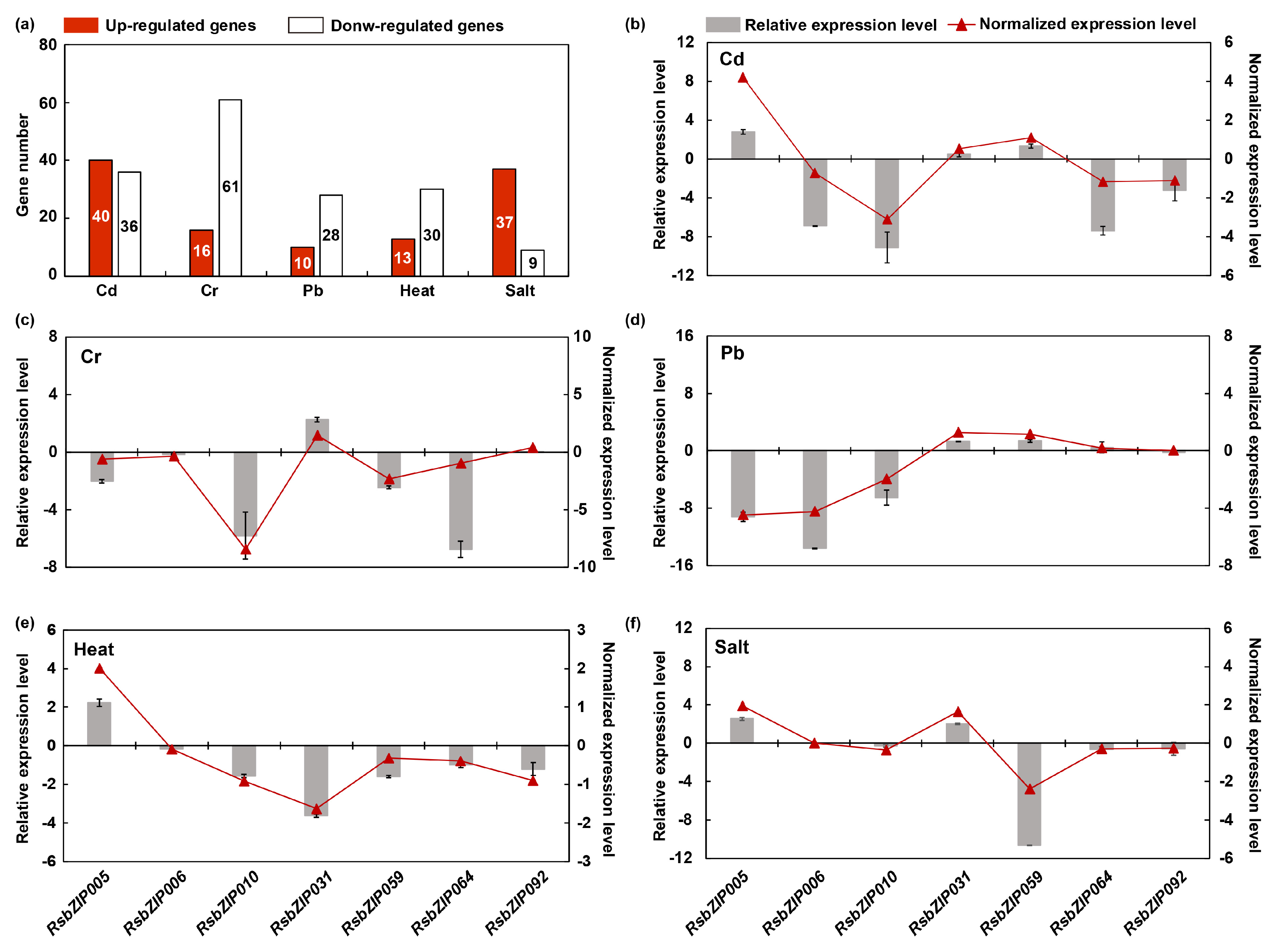
2.5. Potential Roles of RsbZIP Genes in Various Abiotic Stress Responses
2.6. Potential Roles of RsbZIP Genes in Anthocyanin Biosynthetic Pathway
3. Discussion
3.1. Characterization of bZIP Gene Family Members in Radish
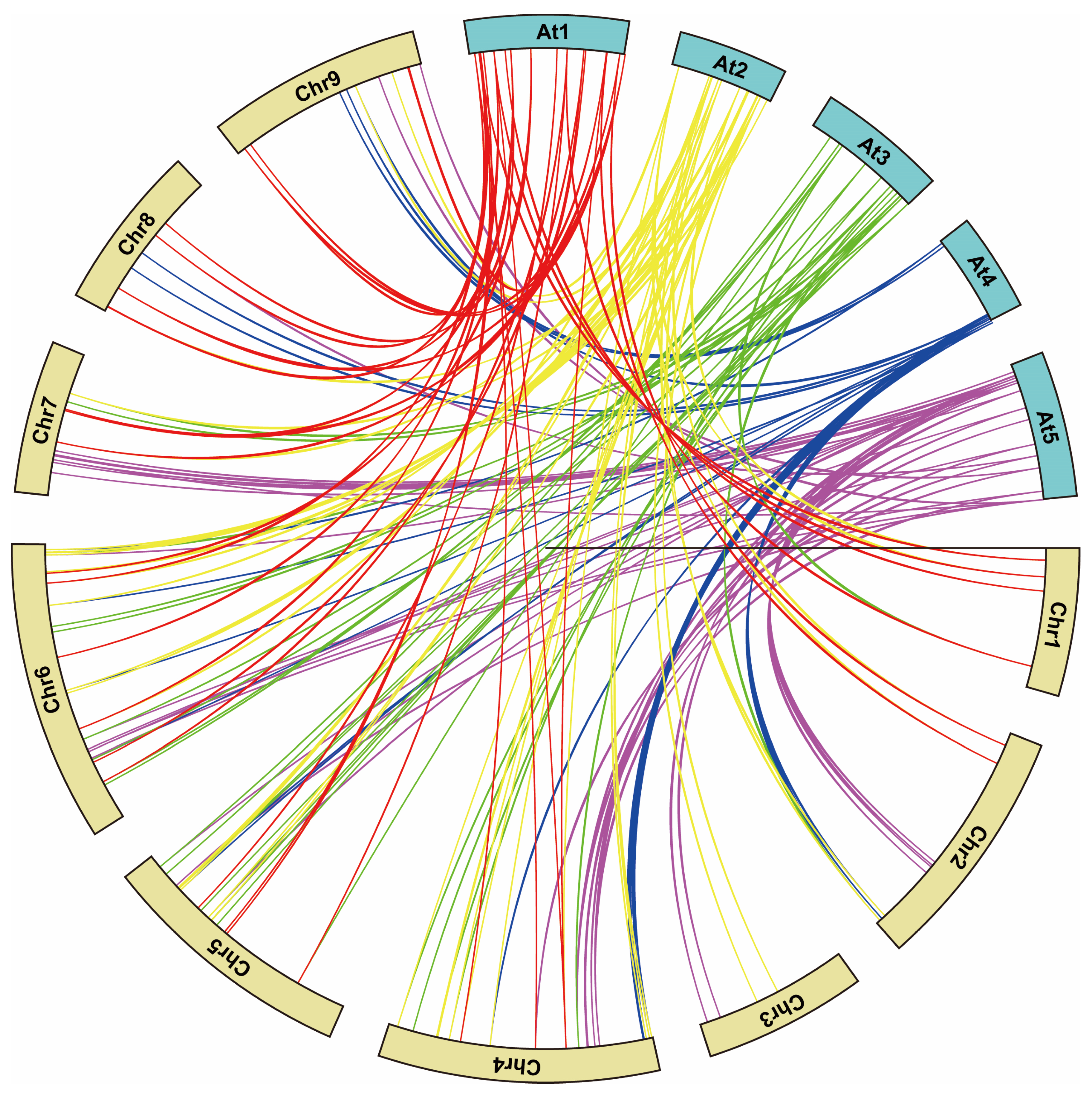
3.2. Collinearity-Orthologues of the bZIP Genes between Radish and Arabidopsis
3.3. Roles of RsbZIP Genes in Different Tissues and Abiotic Stress Responses
3.4. Potential Function of bZIP Genes in Regulation of Anthocyanin Biosynthesis
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of the bZIP Gene Family in Radish
4.2. Synteny Analysis and Chromosomal Localization
4.3. Analyses of Gene Structure, Conserved Motif, Promoter and GO Annotation
4.4. Transcriptome-Based Expression Profiling of RsbZIP Genes
4.5. Plant Materials and Treatments
4.6. Determination of Anthocyanin Content
4.7. RT-qPCR Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Amino acids | aa |
| bZIP | Basic leucine zipper |
| Cd | Cadmium |
| Cr | Chromium |
| Chr(s) | Chromosome(s) |
| DAG | Days after germination |
| GO | Gene ontology |
| HM | Heavy metal |
| HY5 | Elongated hypocotyl 5 |
| HYH | HY5 homolog |
| Pb | Lead |
| Leu | Leucine |
| NJ | Neighbor-Joining |
| RGD | Radish Genome Database |
| RPKM | Reads per kilobase per million reads |
| RT-qPCR | Real-Time Quantitative Polymerase Chain Reaction |
References
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cheng, K.; Wan, L.; Yan, L.; Jiang, H.; Liu, S.; Lei, Y.; Liao, B. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genom. 2015, 16, 1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unel, N.M.; Cetin, F.; Karaca, Y.; Celik, A.Y.; Baloglu, M.C. Comparative identification, characterization, and expression analysis of bZIP gene family members in watermelon and melon genomes. Plant Growth Regul. 2019, 87, 227–243. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Bin, C.; Santo, S.D.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [Green Version]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Guo, R.; Guo, C.; Hou, H.; Wang, X.; Gao, H. Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front. Plant Sci. 2016, 7, 376. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-wide identification and structural analysis of bZIP transcription factor genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.S.; Yang, H.; Ding, L.; Song, Z.T.; Ma, H.; Chang, F.; Liu, J.X. Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 2017, 29, 1007–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.C.; Chi, C.; Jin, L.J.; Zhu, J.; Yu, J.Q.; Zhou, Y.H. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Kitashiba, H.; Li, F.; Hirakawa, H.; Kawanabe, T.; Zou, Z.; Hasegawa, Y.; Tonosaki, K.; Shirasawa, S.; Fukushima, A.; Yokoi, S.; et al. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 2014, 21, 481–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Qiu, Y.; Duan, M.; Wang, J.; Zhang, X.; Wang, H.; Song, J.; Li, X. Identification of anthocyanin biosynthesis related microRNAs in a distinctive Chinese radish (Raphanus sativus L.) by high-throughput sequencing. Mol. Genet. Genom. 2017, 292, 215–229. [Google Scholar] [CrossRef]
- Mitsui, Y.; Shimomura, M.; Komatsu, K.; Namiki, N.; Shibata-Hatta, M.; Imai, M.; Yuichi, K.; Mukai, Y.; Kanamori, H.; Kurita, K.; et al. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci. Rep. 2015, 5, 10835. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wang, Y.; Liu, W.; Wang, J.; Zhu, X.; Zhang, K.; Yu, R.; Wang, R.; Xie, Y.; Zhang, W.; et al. De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant Sci. 2015, 236, 313–323. [Google Scholar] [CrossRef]
- Xie, Y.; Ye, S.; Wang, Y.; Xu, L.; Zhu, X.; Yang, J.; Feng, H.; Yu, R.; Karanja, B.; Gong, Y.; et al. Transcriptome-based gene profiling provides novel insights into the characteristics of radish root response to Cr stress with next-generation sequencing. Front. Plant Sci. 2015, 6, 202. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Chen, Y.; Shen, H.; Gong, Y.; Limera, C.; Liu, L. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to Lead (Pb) stress with next generation sequencing. PLoS ONE 2013, 8, e66539. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Mei, Y.; Xu, L.; Zhu, X.; Wang, Y.; Guo, J.; Liu, L. Genome-wide characterization of differentially expressed genes provides insights into regulatory network of heat stress response in radish (Raphanus sativus L.). Funct. Integr. Genom. 2018, 18, 225–239. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Luo, X.; Zhu, X.; Kinuthia, K.B.; Nie, S.; Feng, H.; Li, C.; Liu, L. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghiurcuta, C.G.; Moret, B.M. Evaluating synteny for improved comparative studies. Bioinformatics 2014, 30, i9–i18. [Google Scholar] [CrossRef]
- Hartmann, L.; Pedrotti, L.; Weiste, C.; Fekete, A.; Schierstaedt, J.; Göttler, J.; Kempa, S.; Krischke, M.; Dietrich, K.; Mueller, M.J.; et al. Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell 2015, 27, 2244–2260. [Google Scholar] [CrossRef]
- Lim, S.H.; Song, J.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Kim, Y.M.; Ha, S.H. Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 2016, 35, 641–653. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Costantini, L.; Malacarne, G.; Lorenzi, S.; Troggio, M.; Mattivi, F.; Moser, C.; Grando, M.S. New candidate genes for the fine regulation of the colour of grapes. J. Exp. Bot. 2015, 66, 4427–4440. [Google Scholar] [CrossRef]
- Ilk, N.; Ding, J.; Ihnatowicz, A.; Koornneef, M.; Matthieu, R. Natural variation for anthocyanin accumulation under high-light and low-temperature stress is attributable to the ENHANCER OF AG-4 2 (HUA2) locus in combination with PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2. New Phytol. 2015, 206, 422–435. [Google Scholar] [CrossRef]
- Rienth, M.; Torregrosa, L.; Luchaire, N.; Chatbanyong, R.; Lecourieux, D.; Kelly, M.T.; Romieu, C. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol. 2014, 14, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, Y.M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.H.; Chung, H.; Jeong, S.; Lim, K.B.; Hwang, Y.J.; Kim, G.B.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. Smart: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sanjari, S.; Shirzadian-Khorramabad, R.; Shobbar, Z.S.; Shahbazi, M. Systematic analysis of NAC transcription factors’ gene family and identification of post-flowering drought stress responsive members in sorghum. Plant Cell Rep. 2019, 38, 361–376. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Wu, J.; Zheng, S.; Cheng, F.; Liu, B.; Liang, J.; Cui, Y.; Wang, X. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp. Chinensis var. purpurea). Mol. Breed. 2015, 35, 113. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, X.; Gong, Y.; Xu, L.; Wang, Y.; Liu, L. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2012, 424, 398–403. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, L.; Xu, L.; Wang, Y.; Tang, M.; Liu, L. Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.). Int. J. Mol. Sci. 2019, 20, 6334. https://doi.org/10.3390/ijms20246334
Fan L, Xu L, Wang Y, Tang M, Liu L. Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.). International Journal of Molecular Sciences. 2019; 20(24):6334. https://doi.org/10.3390/ijms20246334
Chicago/Turabian StyleFan, Lianxue, Liang Xu, Yan Wang, Mingjia Tang, and Liwang Liu. 2019. "Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.)" International Journal of Molecular Sciences 20, no. 24: 6334. https://doi.org/10.3390/ijms20246334




