The Membrane Proximal Domain of TRPV1 and TRPV2 Channels Mediates Protein–Protein Interactions and Lipid Binding In Vitro
Abstract
:1. Introduction
2. Results
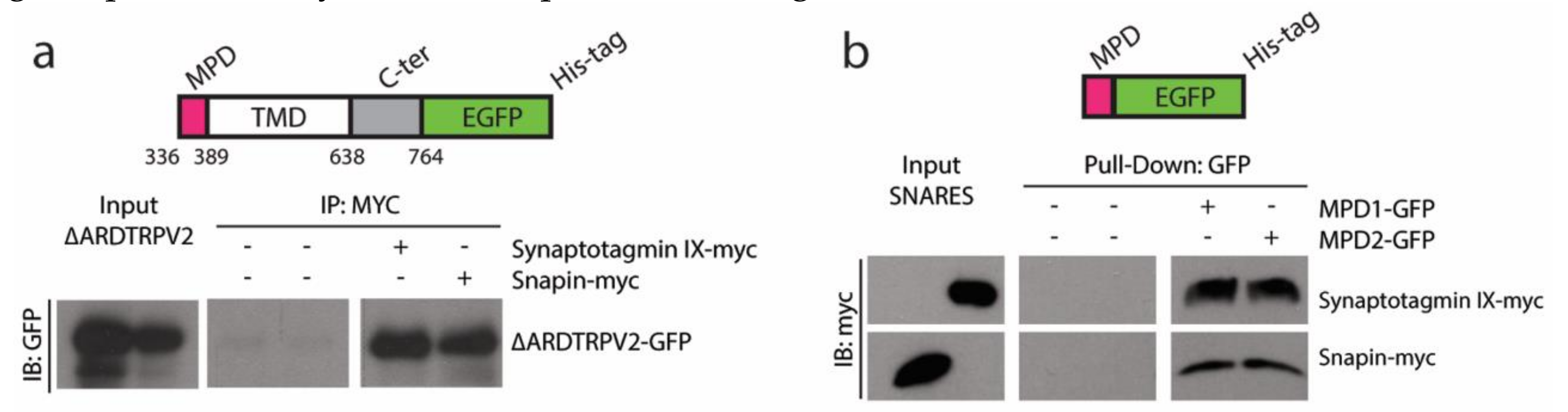
2.1. TRPVs and SNARE-Protein MPD-Mediated Interaction
2.2. MPD-Mediated Lipid Binding
2.3. MPD Oligomerization State
3. Discussion
4. Materials and Methods
4.1. DNA Plasmids and Cloning
4.2. Recombinant Protein Expression and Purification
4.3. Cell Cultures and Transfection
4.4. Immunoblotting
4.5. Co-Immunoprecipitation
4.6. Pull Down
4.7. Lipid Strips
4.8. Tryptophan Quenching
4.9. SAXS Data
4.10. MSA and Tridimensional Representations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MPD | Membrane proximal domain |
| TRP | Transient receptor potential |
| TRPV | Transient receptor potential vanilloid |
| SNARE GFP Stx Syt PLD PA PI PI4P PC PS PG | SNAP (Soluble N-ethylmaleimide-sensitive factor Attachment Protein) REceptor Green fluorescent protein Syntaxins Synaptotagmin Phospholipase D Phosphatidic acid Phosphatidylinositol Phosphatidylinositol 4-phosphate Phosphatidylcholine Phosphatidylserine Phosphatidylglycerol |
References
- Bennett, M.K.; Calakos, N.; Scheller, R.H. Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 1992, 257, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Ilardi, J.M.; Mochida, S.; Sheng, Z.H. Snapin: A SNARE-associated protein implicated in synaptic transmission. Nat. Neurosci. 1999, 2, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.R.; Hanson, P.I.; An, S.; Jahn, R. Ca2+ regulates the interaction between synaptotagmin and syntaxin 1. J. Biol. Chem. 1995, 270, 23667–23671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kim-Miller, M.J.; Fukuda, M.; Kowalchyk, J.A.; Martin, T.F.J. Ca2+-dependent synaptotagmin binding to SNAP-25 is essential for Ca2+-triggered exocytosis. Neuron 2002, 34, 599–611. [Google Scholar] [CrossRef]
- Wiser, O.; Bennett, M.K.; Atlas, D. Functional interaction of syntaxin and SNAP-25 with voltage-sensitive L- and N-type Ca2+ channels. EMBO J. 1996, 15, 4100–4110. [Google Scholar] [CrossRef] [PubMed]
- Ferrandiz-Huertas, C.; Mathivanan, S.; Wolf, C.J.; Devesa, I.; Ferrer-Montiel, A. Trafficking of ThermoTRP Channels. Membranes 2014, 4, 525–564. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Montell, C. Forcing open TRP channels: Mechanical gating as a unifying activation mechanism. Biochem. Biophys. Res. Commun. 2015, 460, 22–25. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.B.; Gray, J.; Gunthorpe, M.J.; Hatcher, J.P.; Davey, P.T.; Overend, P.; Harries, M.H.; Latcham, J.; Clapham, C.; Atkinson, K.; et al. Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature 2000, 405, 183–187. [Google Scholar] [CrossRef] [Green Version]
- Caterina, M.J.; Rosen, T.A.; Tominaga, M.; Brake, A.J.; Julius, D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 1999, 398, 436–441. [Google Scholar] [CrossRef]
- Morenilla-Palao, C.; Planells-Cases, R.; García-Sanz, N.; Ferrer-Montiel, A. Regulated exocytosis contributes to protein kinase C potentiation of vanilloid receptor activity. J. Biol. Chem. 2004, 279, 25665–25672. [Google Scholar] [CrossRef]
- Doñate-Macián, P.; Gómez, A.; Dégano, I.R.; Perálvarez-Marín, A. A TRPV2 interactome-based signature for prognosis in glioblastoma patients. Oncotarget 2018, 9, 18400–18409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lishko, P.V.; Procko, E.; Jin, X.; Phelps, C.B.; Gaudet, R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron 2007, 54, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Cao, E.; Julius, D.; Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Liu, B.; Qin, F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. USA 2011, 108, 11109–11114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doñate-Macián, P.; Perálvarez-Marín, A. Dissecting domain-specific evolutionary pressure profiles of transient receptor potential vanilloid subfamily members 1 to 4. PLoS ONE 2014, 9, e110715. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, D.A.; Violin, J.D.; Newton, A.C.; Tsien, R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 2002, 296, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.; Wang, Y.; Tang, B.L. Protein family review the syntaxins. Genome 2001, 11, reviews3012.1–reviews3012.7. [Google Scholar]
- Fernandez, I.; Ubach, J.; Dulubova, I.; Zhang, X.; Südhof, T.C.; Rizo, J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell 1998, 94, 841–849. [Google Scholar] [CrossRef]
- Lam, A.D.; Tryoen-Toth, P.; Tsai, B.; Vitale, N.; Stuenkel, E.L. SNARE-catalyzed Fusion Events Are Regulated by Syntaxin1A-Lipid Interactions. Mol. Biol. Cell 2008, 19, 485–497. [Google Scholar] [CrossRef]
- Nieto-Posadas, A.; Picazo-juárez, G.; Llorente, I.; Jara-oseguera, A.; Morales-Lázaro, S.; Escalante-Alcalde, D.; Islas, L.D.; Rosenbaum, T. Lysophosphatidic acid directly activates TRPV1 through a C-terminal binding site. Nat. Chem. Biol. 2012, 8, 78–85. [Google Scholar] [CrossRef]
- Shin, J.J.; Loewen, C.J. Putting the pH into phosphatidic acid signaling. BMC Biol. 2011, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.A.; Giudici, A.M.; Renart, M.L.; Molina, M.L.; Montoya, E.; Fernández-Carvajal, A.; Fernández-Ballester, G.; Encinar, J.A.; González-Ros, J.M. Lipid modulation of ion channels through specific binding sites. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1560–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukacs, V.; Thyagarajan, B.; Varnai, P.; Balla, A.; Balla, T.; Rohacs, T. Dual regulation of TRPV1 by phosphoinositides. J. Neurosci. 2007, 27, 7070–7080. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Elias, A.; Mrkonjic, S.; Pardo-Pastor, C.; Inada, H.; Hellmich, U.A.; Rubio-Moscardó, F.; Plata, C.; Gaudet, R.; Vicente, R.; Valverde, M.A. Phosphatidylinositol-4,5-biphosphate-dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proc. Natl. Acad. Sci. USA 2013, 110, 9553–9558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeniou-Meyer, M.; Zabari, N.; Ashery, U.; Chasserot-Golaz, S.; Haeberle, A.M.; Demais, V.; Bailly, Y.; Gottfried, I.; Nakanishi, H.; Neiman, A.M.; et al. Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J. Biol. Chem. 2007, 282, 21746–21757. [Google Scholar] [CrossRef] [PubMed]
- Chernomordik, L.V.; Kozlov, M.M. Protein-Lipid Interplay in Fusion and Fission of Biological Membranes. Annu. Rev. Biochem. 2003, 72, 175–207. [Google Scholar] [CrossRef] [PubMed]
- Hite, R.K.; Butterwick, J.A.; MacKinnon, R. Phosphatidic acid modulation of Kv channel voltage sensor function. Elife 2014, 3, e04366. [Google Scholar] [CrossRef]
- Garcia-Elias, A.; Berna-Erro, A.; Rubio-Moscardo, F.; Pardo-Pastor, C.; Mrkonjić, S.; Sepúlveda, R.V.; Vicente, R.; González-Nilo, F.; Valverde, M.A. Interaction between the Linker, Pre-S1, and TRP Domains Determines Folding, Assembly, and Trafficking of TRPV Channels. Structure 2015, 23, 1404–1413. [Google Scholar] [CrossRef] [Green Version]
- Taberner, F.J.; Fernández-Ballester, G.; Fernández-Carvajal, A.; Ferrer-Montiel, A. TRP channels interaction with lipids and its implications in disease. Biochim. Biophys. Acta Biomembr. 2015, 1848, 1818–1827. [Google Scholar] [CrossRef] [Green Version]
- Ciardo, M.G.; Ferrer-Montiel, A. Lipids as central modulators of sensory TRP channels. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1615–1628. [Google Scholar] [CrossRef]
- SAXSIT. Available online: https://www.slri.or.th/th/index.php/beamline/bl13w.html? (accessed on 04 February 2019).
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Svergun, D. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999, 76, 2879–2886. [Google Scholar] [CrossRef]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; Dicuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucl. Acids Res. 2008, 36, D13–D21. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doñate-Macián, P.; Álvarez-Marimon, E.; Sepulcre, F.; Vázquez-Ibar, J.L.; Perálvarez-Marín, A. The Membrane Proximal Domain of TRPV1 and TRPV2 Channels Mediates Protein–Protein Interactions and Lipid Binding In Vitro. Int. J. Mol. Sci. 2019, 20, 682. https://doi.org/10.3390/ijms20030682
Doñate-Macián P, Álvarez-Marimon E, Sepulcre F, Vázquez-Ibar JL, Perálvarez-Marín A. The Membrane Proximal Domain of TRPV1 and TRPV2 Channels Mediates Protein–Protein Interactions and Lipid Binding In Vitro. International Journal of Molecular Sciences. 2019; 20(3):682. https://doi.org/10.3390/ijms20030682
Chicago/Turabian StyleDoñate-Macián, Pau, Elena Álvarez-Marimon, Francesc Sepulcre, José Luis Vázquez-Ibar, and Alex Perálvarez-Marín. 2019. "The Membrane Proximal Domain of TRPV1 and TRPV2 Channels Mediates Protein–Protein Interactions and Lipid Binding In Vitro" International Journal of Molecular Sciences 20, no. 3: 682. https://doi.org/10.3390/ijms20030682




