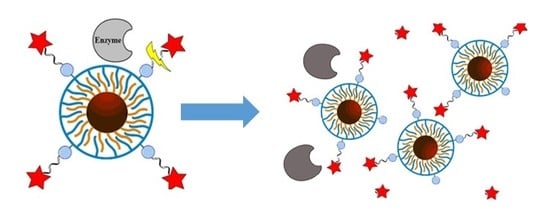
Investigating Possible Enzymatic Degradation on Polymer Shells around Inorganic Nanoparticles
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mazuel, F.O.; Espinosa, A.; Luciani, N.; Reffay, M.; Borgne, R.M.L.; Motte, L.; Desboeufs, K.; Michel, A.; Pellegrino, T.; Lalatonne, Y.; et al. Massive Intracellular Biodegradation of Iron Oxide Nanoparticles Evidenced Magnetically at Single-Endosome and Tissue Levels. ACS Nano 2016, 10, 7627–7638. [Google Scholar] [CrossRef] [PubMed]
- Kolosnjaj-Tabi, J.; Javed, Y.; Lartigue, L.; Volatron, J.; Elgrabli, D.; Marangon, I.; Pugliese, G.; Caron, B.; Figuerola, A.; Luciani, N.; et al. The One Year Fate of Iron Oxide Coated Gold Nanoparticles in Mice. ACS Nano 2015, 9, 7925–7939. [Google Scholar] [CrossRef]
- Feliu, N.; Docter, D.; Heine, M.; Pino, P.D.; Ashraf, S.; Kolosnjaj-Tabi, J.; Macchiarini, P.; Nielsen, P.; Alloyeau, D.; Gazeau, F.; et al. In vivo degradation and the fate of inorganic nanoparticles. Chem. Soc. Rev. 2016, 45, 2440–2457. [Google Scholar] [CrossRef]
- Soenen, S.J.H.; Himmelreich, U.; Nuytten, N.; Pisanic Ii, T.R.; Ferrari, A.; De Cuyper, M. Intracellular Nanoparticle Coating Stability Determines Nanoparticle Diagnostics Efficacy and Cell Functionality. Small 2010, 6, 2136–2145. [Google Scholar] [CrossRef] [PubMed]
- Rivera Gil, P.; Jimenez de Aberasturi, D.; Wulf, V.; Pelaz, B.; del Pino, P.; Zhao, Y.; de la Fuente, J.; Ruiz de Larramendi, I.; Rojo, T.; Liang, X.-J.; et al. The Challenge to Relate the Physicochemical Properties of Colloidal Nanoparticles to Their Cytotoxicity. Acc. Chem. Res. 2013, 46, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Spector, L.B. Covalent enzyme-substrate intermediates in transferase reactions. Bioorg. Chem. 1973, 2, 311–321. [Google Scholar] [CrossRef]
- Chanana, M.; Rivera Gil, P.; Correa-Duarte, M.A.; Parak, W.J.; Liz-Marzán, L.M. Physicochemical properties of protein-coated gold nanoparticles in biological fluids and cells before and after proteolytic digestion. Angew. Chem. Int. Ed. 2013, 52, 4179–4183. [Google Scholar] [CrossRef]
- Kreyling, W.G.; Abdelmonem, A.M.; Ali, Z.; Alves, F.; Geiser, M.; Haberl, N.; Hartmann, R.; Hirn, S.; de Aberasturi, D.J.; Kantner, K.; et al. In vivo integrity of polymer-coated gold nanoparticles. Nat. Nanotechnol. 2015, 10, 619–623. [Google Scholar] [CrossRef]
- Llop, J.; Jiang, P.; Marradi, M.; Gomez-Vallejo, V.; Echeverria, M.; Yu, S.; Puigivila, M.; Baz, Z.; Szczupak, B.; Perez-Campana, C.; et al. Visualisation of dual radiolabelled poly(lactide-co-glycolide) nanoparticle degradation in vivo using energy-discriminant SPECT. J. Mater. Chem. B 2015, 3, 6293–6300. [Google Scholar] [CrossRef] [Green Version]
- Sée, V.; Free, P.; Cesbron, Y.; Nativo, P.; Shaheen, U.; Rigden, D.; Spiller, D.G.; Fernig, D.G.; White, M.R.H.; Prior, I.A.; et al. Cathepsin L digestion of nanobioconjugates upon endocytosis. ACS Nano 2009, 3, 2461–2468. [Google Scholar] [CrossRef]
- Lunov, O.; Syrovets, T.; Rocker, C.; Tron, K.; Nienhaus, G.U.; Rasche, V.; Mailander, V.; Landfester, K.; Simmet, T. Lysosomal degradation of the carboxydextran shell of coated superparamagnetic iron oxide nanoparticles and the fate of professional phagocytes. Biomaterials 2010, 31, 9015–9022. [Google Scholar] [CrossRef] [PubMed]
- Kohler, N.; Sun, C.; Wang, J.; Zhang, M. Methotrexate-modified superparamagnetic nanoparticles and their intracellular uptake into human cancer cells. Langmuir 2005, 21, 8858–8864. [Google Scholar] [CrossRef]
- Chen, H.W.; Zou, P.; Connarn, J.; Paholak, H.; Sun, D.X. Intracellular dissociation of a polymer coating from nanoparticles. Nano Res. 2012, 5, 815–825. [Google Scholar] [CrossRef]
- Akagi, T.; Higashi, M.; Kaneko, T.; Kida, T.; Akashi, M. Hydrolytic and enzymatic degradation of nanoparticles based on amphiphilic poly(gamma-glutamic acid)-graft-l-phenylalanine copolymers. Biomacromolecules 2006, 7, 297–303. [Google Scholar] [CrossRef]
- De la Rica, R.; Aili, D.; Stevens, M.M. Enzyme-responsive nanoparticles for drug release and diagnostics. Adv. Drug Del. Rev. 2012, 64, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Qiu, N.S.; Liu, X.R.; Zhong, Y.; Zhou, Z.X.; Piao, Y.; Miao, L.; Zhang, Q.Z.; Tang, J.B.; Huang, L.; Shen, Y.Q. Esterase-Activated Charge-Reversal Polymer for Fibroblast-Exempt Cancer Gene Therapy. Adv. Mater. 2016, 28, 10613–10622. [Google Scholar] [CrossRef]
- Huang, S.X.; Shao, K.; Kuang, Y.Y.; Liu, Y.; Li, J.F.; An, S.; Guo, Y.B.; Ma, H.J.; He, X.; Jiang, C. Tumor targeting and microenvironment-responsive nanoparticles for gene delivery. Biomaterials 2013, 34, 5294–5302. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.E.; Geng, J.; Pu, F.; Yang, X.J.; Ren, J.S.; Qu, X.G. Polyvalent Nucleic Acid/Mesoporous Silica Nanoparticle Conjugates: Dual Stimuli-Responsive Vehicles for Intracellular Drug Delivery. Angew. Chem.-Int. Ed. 2011, 50, 882–886. [Google Scholar] [CrossRef]
- Hühn, J.; Carrillo-Carrion, C.; Soliman, M.G.; Pfeiffer, C.; Valdeperez, D.; Masood, A.; Chakraborty, I.; Zhu, L.; Gallego, M.; Zhao, Y.; et al. Selected Standard Protocols for the Synthesis, Phase Transfer, and Characterization of Inorganic Colloidal Nanoparticles. Chem. Mater. 2017, 29, 399–461. [Google Scholar] [CrossRef]
- Colombo, M.; Fiandra, L.; Alessio, G.; Mazzucchelli, S.; Nebuloni, M.; Palma, C.D.; Kantner, K.; Pelaz, B.; Rotem, R.; Corsi, F.; et al. Tumour homing and therapeutic effect of colloidal nanoparticles depend on the number of attached antibodies. Nat. Commun. 2016, 7, 13818. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.H.; Iribarren, P.; Zhang, N.; Zhou, Y.; Gong, W.H.; Cho, E.H.; Lockett, S.; Chertov, O.; Bednar, F.; Rogers, T.J.; et al. Identification of neutrophil granule protein cathepsin G as a novel chemotactic agonist for the G protein-coupled formyl peptide receptor. J. Immunol. 2004, 173, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Corrias, A.; Mountjoy, G.; Loche, D.; Puntes, V.; Falqui, A.; Zanella, M.; Parak, W.J.; Casula, M.F. Identifying Spinel Phases in Nearly Monodisperse Iron Oxide Colloidal Nanocrystal. J. Phys. Chem. C 2009, 113, 18667–18675. [Google Scholar] [CrossRef] [Green Version]
- Cozzoli, P.D.; Snoeck, E.; Garcia, M.A.; Giannini, C.; Guagliardi, A.; Cervellino, A.; Gozzo, F.; Hernando, A.; Achterhold, K.; Ciobanu, N.; et al. Colloidal synthesis and characterization of tetrapod-shaped magnetic nanocrystals. Nano Lett. 2006, 6, 1966–1972. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, L.; Alvarez-Puebla, R.A.; Pazos-Perez, N. Surface Modifications of Nanoparticles for Stability in Biological Fluids. Materials 2018, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Pazos-Perez, N.; Rodriguez-Gonzalez, B.; Hilgendorff, M.; Giersig, M.; Liz-Marzan, L.M. Gold encapsulation of star-shaped FePt nanoparticles. J. Mater. Chem. 2010, 20, 61–64. [Google Scholar] [CrossRef]
- Martínez-Boubeta, C.; Simeonidis, K.; Angelakeris, M.; Pazos-Pérez, N.; Giersig, M.; Delimitis, A.; Nalbandian, L.; Alexandrakis, V.; Niarchos, D. Critical radius for exchange bias in naturally oxidized Fe nanoparticles. Phys. Rev. B 2006, 74, 054430. [Google Scholar] [CrossRef]
- Pazos-Perez, N.; Gao, Y.; Hilgendorff, M.; Irsen, S.; Perez-Juste, J.; Spasova, M.; Farle, M.; Liz-Marzan, L.M.; Giersig, M. Magnetic-noble metal nanocomposites with morphology-dependent optical response. Chem. Mater. 2007, 19, 4415–4422. [Google Scholar] [CrossRef]
- Lin, C.-A.J.; Sperling, R.A.; Li, J.K.; Yang, T.-Y.; Li, P.-Y.; Zanella, M.; Chang, W.H.; Parak, W.J. Design of an Amphiphilic Polymer for Nanoparticle Coating and Functionalization. Small 2008, 4, 334–341. [Google Scholar] [CrossRef]
- Liang, L.Y.; Astruc, D. The copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) “click” reaction and its applications. An overview. Coord. Chem. Rev. 2011, 255, 2933–2945. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Snyder, S.A.; Montagnon, T.; Vassilikogiannakis, G. The Diels-Alder reaction in total synthesis. Angew. Chem.-Int. Ed. 2002, 41, 1668–1698. [Google Scholar] [CrossRef]
- Sperling, R.A.; Liedl, T.; Duhr, S.; Kudera, S.; Zanella, M.; Lin, C.-A.J.; Chang, W.H.; Braun, D.; Parak, W.J. Size Determination of (Bio-) Conjugated Water-Soluble Colloidal Nanoparticles: A Comparison of Different Techniques. J. Phys. Chem. C 2007, 111, 11552–11559. [Google Scholar] [CrossRef]
- Yakovlev, A.V.; Zhang, F.; Zulqurnain, A.; Azhar-Zahoor, A.; Luccardini, C.; Gaillard, S.; Mallet, J.M.; Tauc, P.; Brochon, J.C.; Parak, W.J.; et al. Wrapping Nanocrystals with an Amphiphilic Polymer Preloaded with Fixed Amounts of Fluorophore Generates FRET-Based Nanoprobes with a Controlled Donor/Acceptor Ratio. Langmuir 2009, 25, 3232–3239. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, U.; Jimenez de Aberasturi, D.; Vazquez-Gonzalez, M.; Carrillo-Carrion, C.; Niebling, T.; Parak, W.J.; Heimbrodt, W. Determining the exact number of dye molecules attached to colloidal CdSe/ZnS quantum dots in Förster resonant energy transfer assemblies. J. Appl. Phys. 2015, 117, 024701. [Google Scholar] [CrossRef]
- Scheele, G.; Bartelt, D.; Bieger, W. Characterization of human exocrine pancreatic proteins by two-dimensional isoelectric focusing/sodium dodecyl sulfate gel electrophoresis. Gastroenterology 1981, 80, 461–473. [Google Scholar] [PubMed]
- Cottrell, G.S.; Amadesi, S.; Grady, E.F.; Bunnett, N.W. Trypsin IV, a novel agonist of protease-activated receptors 2 and 4. J. Biol. Chem. 2004, 279, 13532–13539. [Google Scholar] [CrossRef]
- Olsen, J.V.; Ong, S.E.; Mann, M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Burster, T.; Macmillan, H.; Hou, T.E.Y.; Boehm, B.O.; Mellins, E.D. Cathepsin G: Roles in antigen presentation and beyond. Mol. Immunol. 2010, 47, 658–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everse, J.; Kaplan, N.O. Lactate Dehydrogenases—Structure and Function. Adv. Enzymol. Relat. Areas Mol. Biol. 1973, 37, 61–133. [Google Scholar]
- Cooper, A.J.L. Glutamate-aspartate transaminase. Methods Enzymol. 1985, 113, 66–69. [Google Scholar]
- Kohler, E.; Seville, M.; Jager, J.; Fotheringham, I.; Hunter, M.; Edwards, M.; Jansonius, J.N.; Kirschner, K. Significant improvement to the catalytic properties of aspartate-aminotransferase—Role of hydrophobic and charged residues in the substrate-binding pocket. Biochemistry 1994, 33, 90–97. [Google Scholar] [CrossRef]
- Saenger, W. Proteinase K; Elsevier Science Bv: Amsterdam, The Netherlands, 2013; pp. 3240–3242. [Google Scholar]
- Van der Valk, J.; Bieback, K.; Buta, C.; Cochrane, B.; Dirks, W.G.; Fu, J.N.; Hickman, J.J.; Hohensee, C.; Kolar, R.; Liebsch, M.; et al. Fetal Bovine Serum (FBS): Past—Present—Future. ALTEX-Altern. Anim. Exp. 2018, 35, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.G.; Marwood, R.M.; Parsons, A.E.; Parsons, R.B. The effect of foetal bovine serum supplementation upon the lactate dehydrogenase cytotoxicity assay: Important considerations for in vitro toxicity analysis. Toxicol. In Vitro 2015, 30, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Nakka, S.; Guruprasad, L.; Samanta, A. Interaction of Bovine Serum Albumin with Dipolar Molecules: Fluorescence and Molecular Docking Studies. J. Phys. Chem. B 2009, 113, 2143–2150. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. Camb. Philos. Soc. 1953, 28, 416–436. [Google Scholar] [CrossRef]
- Menefee, A.L.; Zeczycki, T.N. Nearly 50 years in the making: Defining the catalytic mechanism of the multifunctional enzyme, pyruvate carboxylase. FEBS J. 2014, 281, 1333–1354. [Google Scholar] [CrossRef] [PubMed]
- Hult, K.; Berglund, P. Enzyme promiscuity: Mechanism and applications. Trends Biotechnol. 2007, 25, 231–238. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Pelaz, B.; Chakraborty, I.; Parak, W.J. Investigating Possible Enzymatic Degradation on Polymer Shells around Inorganic Nanoparticles. Int. J. Mol. Sci. 2019, 20, 935. https://doi.org/10.3390/ijms20040935
Zhu L, Pelaz B, Chakraborty I, Parak WJ. Investigating Possible Enzymatic Degradation on Polymer Shells around Inorganic Nanoparticles. International Journal of Molecular Sciences. 2019; 20(4):935. https://doi.org/10.3390/ijms20040935
Chicago/Turabian StyleZhu, Lin, Beatriz Pelaz, Indranath Chakraborty, and Wolfgang J. Parak. 2019. "Investigating Possible Enzymatic Degradation on Polymer Shells around Inorganic Nanoparticles" International Journal of Molecular Sciences 20, no. 4: 935. https://doi.org/10.3390/ijms20040935