Development of the VIGS System in the Dioecious Plant Silene latifolia
Abstract
:1. Introduction
2. Results and Discussion
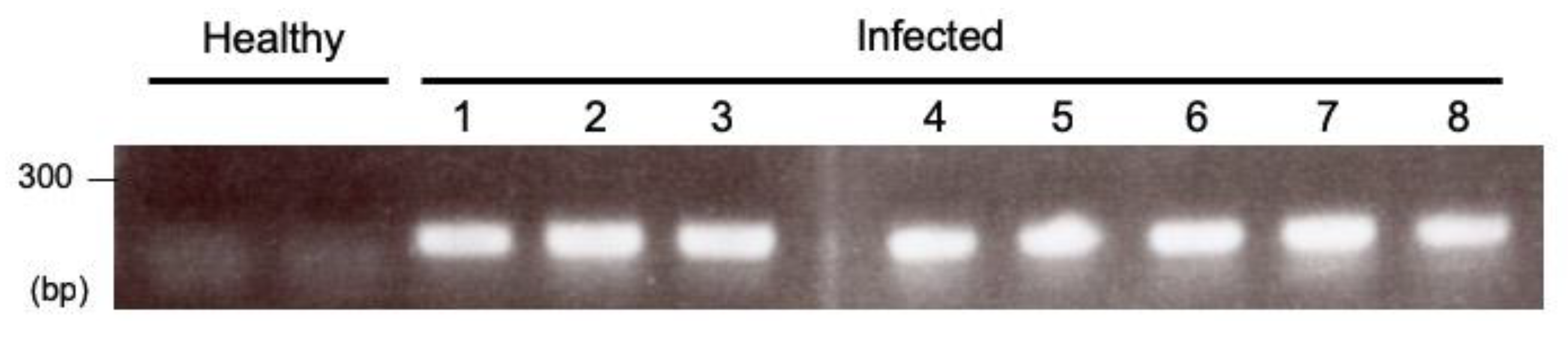
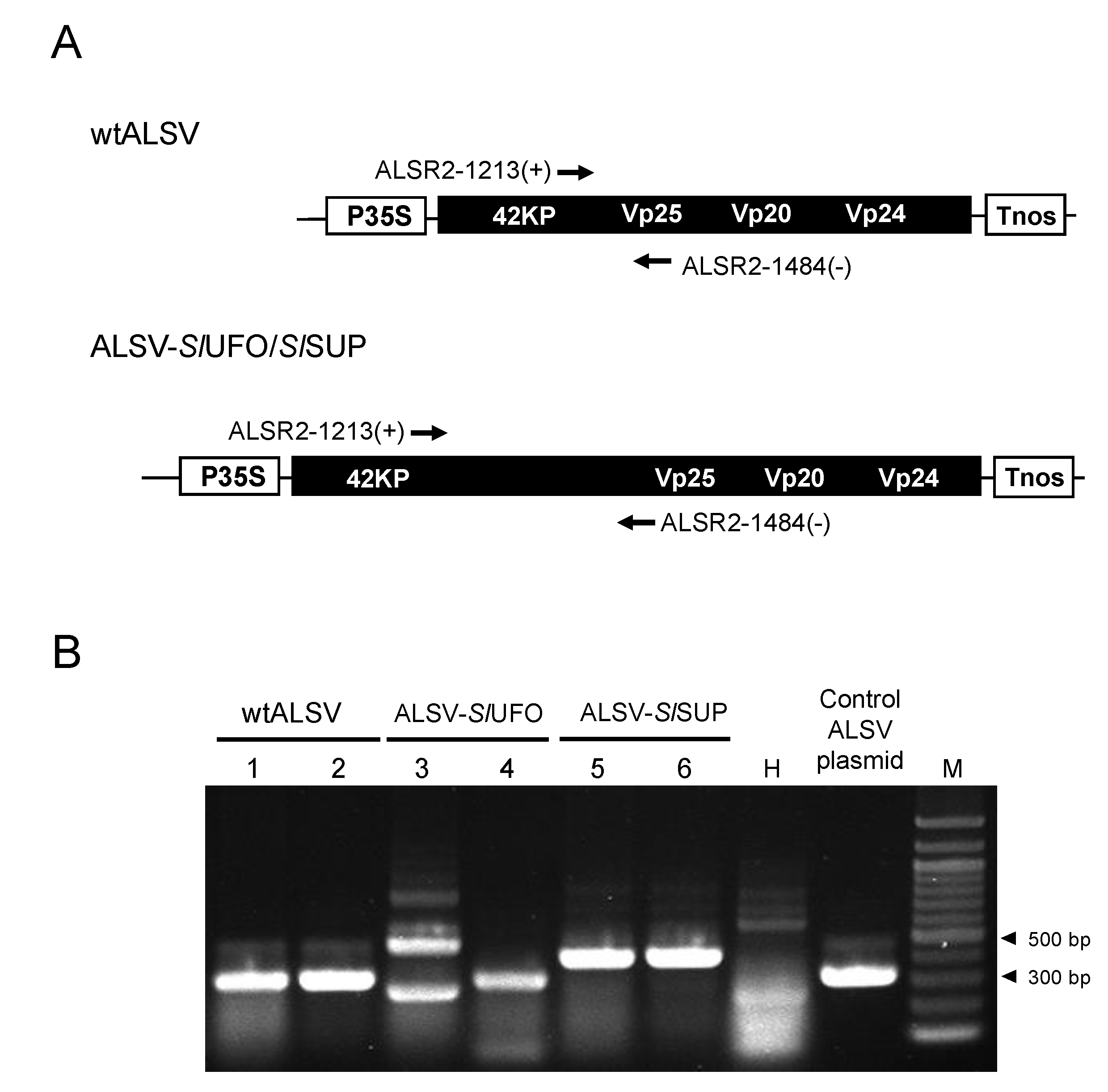
2.1. Optimization of an Inoculation Method for Delivering Apple Latent Spherical Virus (ALSV) to S. latifolia Plants
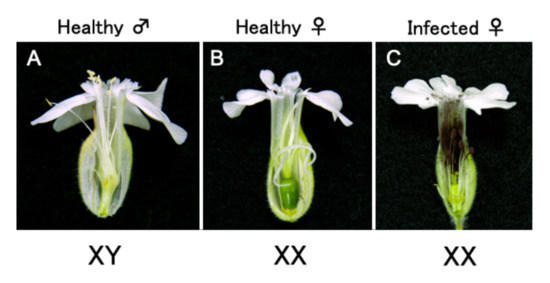
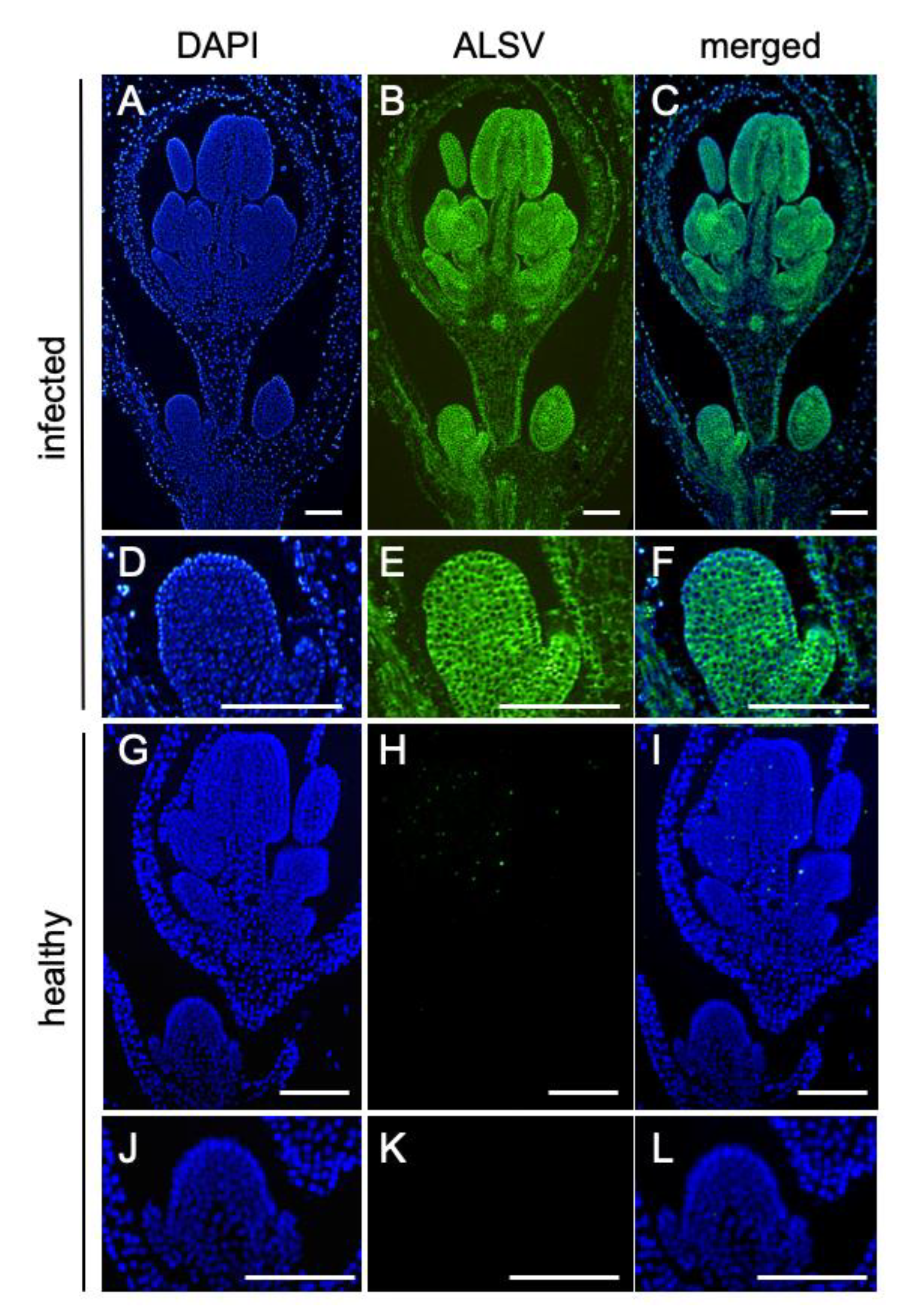
2.2. ALSV Migrates to Flowers in S. latifolia Plants
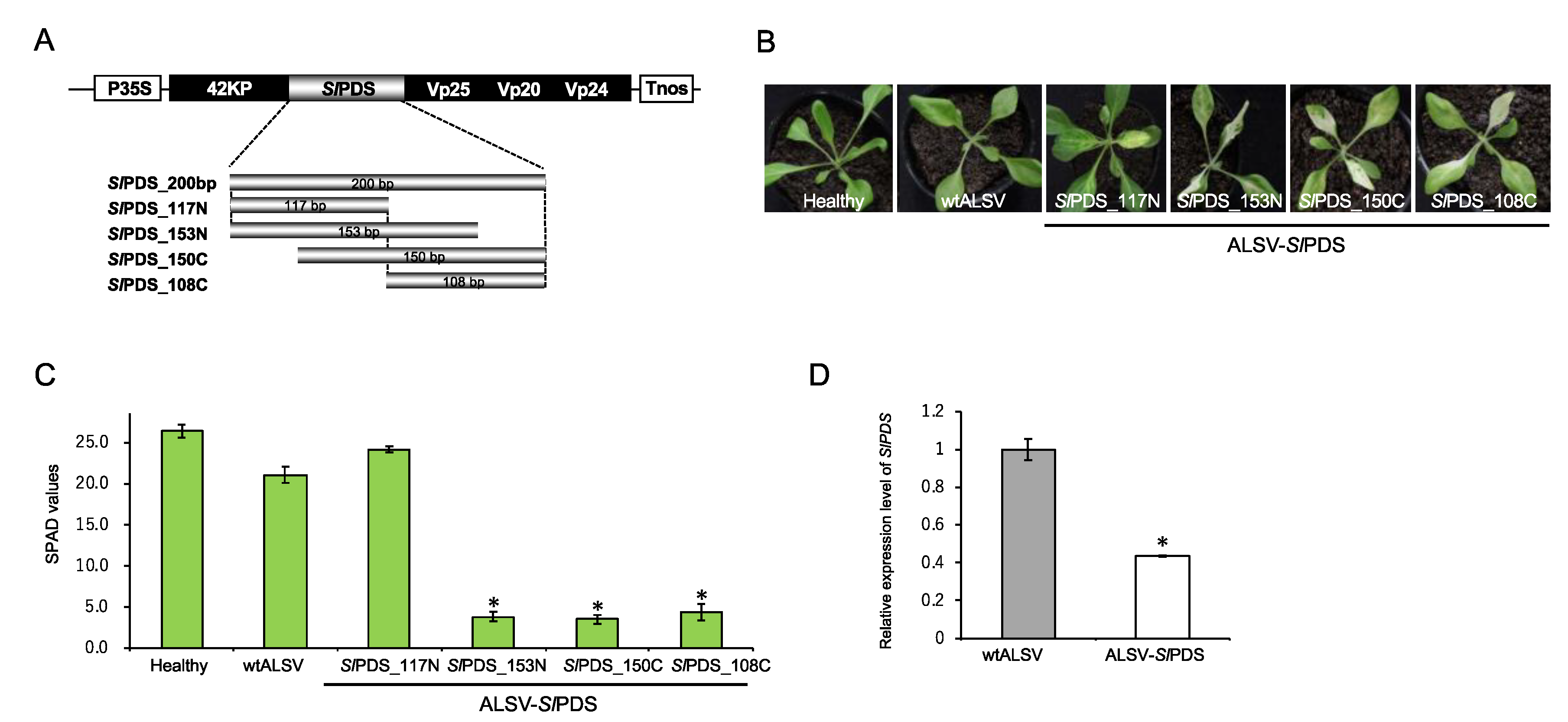
2.3. Evaluation of Virus-Induced Gene Silencing (VIGS)-Mediated Gene Knockdown in S. latifolia Plants
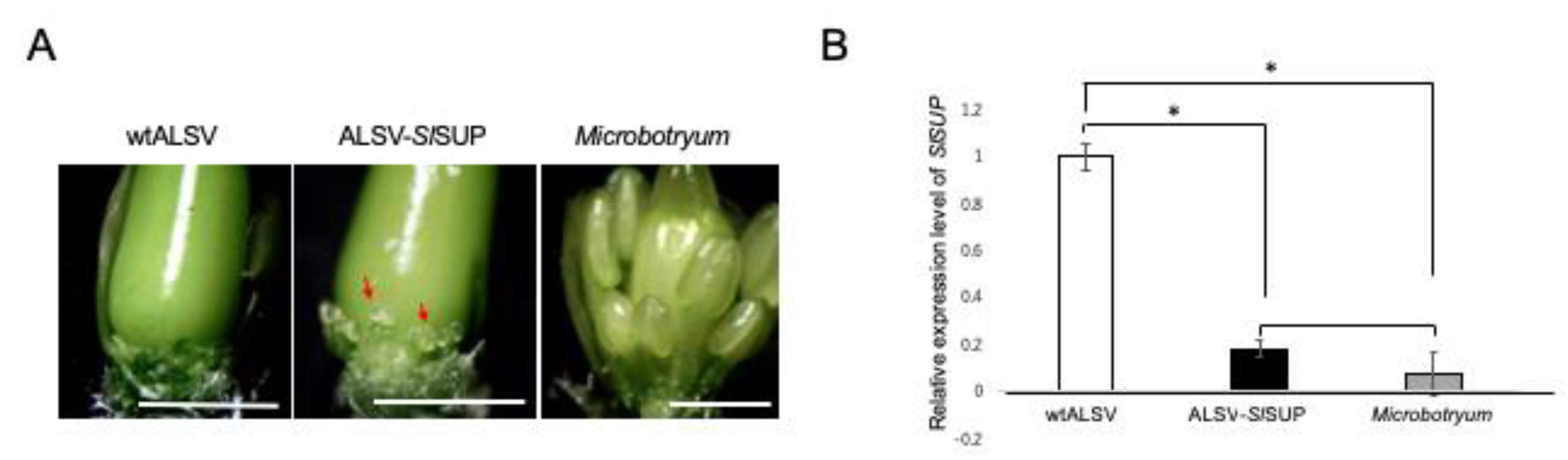
2.4. VIGS-Mediated Gene Knockdown in the Flowers of S. latifolia Plants
3. Materials and Methods
3.1. Plant and Viral Materials, and Agroinfiltration
3.2. Inoculation of ALSV onto S. latifolia Plants
3.3. RNA Extraction and Reverse Transcription Polymerase Chain Reaction (RT-PCR) for Checking ALSV Infection
3.4. In Situ Hybridization Analysis to Detect Virus Invasion into the Floral Meristems
3.5. Construction of ALSV Vectors
3.6. RT-PCR and Quantitative RT-PCR (RT-qPCR)
3.7. Inoculation of Microbotryum onto S. latifolia Plants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Filatov, D.A. The two “rules of speciation” in species with young sex chromosomes. Mol. Ecol. 2018, 27, 3799–3810. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D. Plant sex chromosomes. Annu. Rev. Plant Biol. 2016, 67, 397–420. [Google Scholar] [CrossRef] [PubMed]
- Zemp, N.; Widmer, A.; Charlesworth, D. Has adaptation occurred in males and females since separate sexes evolved in the plant Silene latifolia? Proc. Biol. Sci. 2018, 25, 285. [Google Scholar]
- Pannell, J.R. Plant sex determination. Curr. Biol. 2017, 27, R191–R197. [Google Scholar] [CrossRef] [PubMed]
- Muyle, A.; Zemp, N.; Fruchard, C.; Cegan, R.; Vrana, J.; Deschamps, C.; Tavares, R.; Hobza, R.; Picard, F.; Widmer, A.; et al. Genomic imprinting mediates dosage compensation in a young plant XY system. Nat. Plants 2018, 4, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Lorenzo, J.L.; Hobza, R.; Vyskot, B. DNA methylation and genetic degeneration of the Y chromosome in the dioecious plant Silene latifolia. BMC Genom. 2018, 19, 540. [Google Scholar] [CrossRef] [PubMed]
- Westergaard, M. The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 1958, 9, 217–281. [Google Scholar] [PubMed]
- Uchida, W.; Matsunaga, S.; Sugiyama, R.; Kazama, Y.; Kawano, S. Morphological development of anthers induced by the dimorphic smut fungus Microbotryum violaceum in female flowers of the dioecious plant Silene latifolia. Planta 2003, 218, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, K.B. Sex chromosomes in plants. Nature 1923, 112, 687–688. [Google Scholar] [CrossRef]
- Fujita, N.; Torii, C.; Ishii, K.; Aonuma, W.; Shimizu, Y.; Kazama, Y.; Abe, T.; Kawano, S. Narrowing down the mapping of plant sex-determination regions using new Y-chromosome-specific markers and heavy-ion beam irradiation-induced Y-deletion mutants in Silene latifolia. G3 2012, 2, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Ishii, K.; Aonuma, W.; Ikeda, T.; Kawamoto, H.; Koizumi, A.; Filatov, D.A.; Chibalina, M.; Bergero, R.; Charlesworth, D.; et al. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 2016, 6, 18917. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.S.; Chen, Z.; Rouchka, E.C.; Schultz, D.J.; Cuomo, C.A.; Perlin, M.H. Pas de deux: An intricate dance of anther smut and its host. G3 2018, 8, 505–518. [Google Scholar]
- Hudzieczek, V.; Cegan, R.; Cermak, T.; Bacovska, N.; Machalkova, Z.; Dolezal, K.; Plihalova, L.; Voytas, D.; Hobza, R.; Vyskot, B. Agrobacterium rhizogenes-mediated transformation of a dioecious plant model Silene latifolia. New Biotechnol. 2019, 48, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Minato, N.; Komatsu, K.; Okano, Y.; Maejima, K.; Ozeki, J.; Senshu, H.; Takahashi, S.; Yamaji, Y.; Namba, S. Efficient foreign gene expression in planta using a plantago asiatica mosaic virus-based vector achieved by the strong RNA-silencing suppressor activity of TGBp1. Arch. Virol. 2014, 159, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Schwach, F.; Vaistij, F.E.; Jones, L.; Baulcombe, D.C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005, 138, 1842–1852. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.F.; Huang, Y.H.; Lin, C.P.; Liu, L.Y.D.; Hong, S.F.; Yang, C.Y.; Lo, H.F.; Tseng, T.Y.; Chen, W.Y.; Lin, S.S. Development of a mild viral expression system for gain-of-function study of phytoplasma effector in planta. PLoS ONE 2015, 10, e0130139. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sasaki, M.; Isogai, M.; Yoshikawa, N. Stable expression of foreign proteins in herbaceous and apple plants using Apple latent spherical virus RNA2 vectors. Arch. Virol. 2004, 149, 1541–1558. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A.; Yaegashi, H.; Yamagishi, N.; Takahashi, T.; Isogai, M.; et al. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 2009, 386, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Kasajima, I.; Ito, M.; Yamagishi, N.; Yoshikawa, N. Apple latent spherical virus (ALSV) vector as a tool for reverse genetic studies and non-transgenic breeding of a variety of crops. In Plant Epigenetics, RNA Technologies; Rajewsky, N., Jurga, S., Barciszewski, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 513–536. ISBN 978-3-319-55519-5. [Google Scholar]
- Kung, Y.J.; Lin, P.C.; Yeh, S.D.; Hong, S.F.; Chua, N.H.; Liu, L.Y. Genetic analyses of the FRNK motif function of Turnip mosaic virus uncover multiple and potentially interactive pathways of cross-protection. Mol. Plant Microbe Interact. 2014, 27, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.S.; Langenberg, W.G. Distribution of barley stripe mosaic-virus protein in infected wheat root and shoot tips. J. Gen. Virol. 1984, 65, 2217–2224. [Google Scholar] [CrossRef]
- Sasaki, S.; Yamagishi, N.; Yoshikawa, N. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Plant Methods 2011, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, M.H.; Donsn, J.; Della-Cioppa, G.; Harvey, D.; Hanley, K.; Grill, L.K. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Chae, E.; Tan, Q.K.; Hill, T.A.; Irish, V.F. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 2008, 135, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.F.; Sussex, I.M. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 1990, 2, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.A.; Haughn, G.W. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 1991, 3, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef]
- Bowman, J.L.; Alvarez, J.; Weigel, D.; Meyerowitz, E.M.; Smyth, D.R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 1993, 119, 721–743. [Google Scholar]
- Roth, O.; Alvarez, J.P.; Levy, M.; Bowman, J.L.; Ori, N.; Shani, E. The KNOXI Transcription Factor SHOOT MERISTEMLESS Regulates Floral Fate in Arabidopsis. Plant Cell 2018, 30, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Fujiwara, M.T.; Koizumi, A.; Nishihara, K.; Nishiyama, R.; Kifune, E.; Abe, T.; Kawano, S. A SUPERMAN-like gene is exclusively expressed in female flowers of the dioecious plant Silene latifolia. Plant Cell Physiol. 2009, 50, 1127–1141. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Yamaguchi, H.; Aida, R.; Shikata, M.; Abe, T.; Otsubo, N. Mutation in Torenia fournieri Lind. UFO homolog confers loss of TfLFY interaction and results in a petal to sepal transformation. Plant J. 2012, 71, 1002–1014. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Komatsu, K.; Kagiwada, S.; Ozeki, J.; Mori, T.; Hirata, H.; Yamaji, Y.; Ugaki, M.; Namba, S. The efficiency of interference of Potato virus X infection depends on the target gene. Virus Res. 2006, 116, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, T.; Kawamoto, K. Preparation of thin frozen sections from nonfixed and undecalcified hard tissues using Kawamoto’s film method (2012). Methods Mol. Biol. 2014, 1130, 149–164. [Google Scholar] [PubMed]
- Kazama, Y.; Koizumi, A.; Uchida, W.; Ageez, A.; Kawano, S. Expression of the floral B-function gene SLM2 in female flowers of Silene latifolia infected with the smut fungus Microbotryum violaceum. Plant Cell Physiol. 2005, 46, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Höfgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef] [PubMed]
- Zemp, N.; Minder, A.; Widmer, A. Identification of internal reference genes for gene expression normalization between the two sexes in dioecious white Campion. PLoS ONE 2014, 9, e92893. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Aonuma, W.; Shimizu, Y.; Yamanaka, K.; Hirata, A.; Hood, M.E.; Kawano, S. A petalless flower caused by a Microbotryum violaceum mutant. Int. J. Plant Sci. 2012, 173, 464–473. [Google Scholar] [CrossRef]
- Chujo, T.; Yoshikawa, M.; Ariga, H.; Endo, M.; Toki, S.; Ishibashi, K. A removable virus vector suitable for plant genome editing. Plant J. 2017, 91, 558–561. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujita, N.; Kazama, Y.; Yamagishi, N.; Watanabe, K.; Ando, S.; Tsuji, H.; Kawano, S.; Yoshikawa, N.; Komatsu, K. Development of the VIGS System in the Dioecious Plant Silene latifolia. Int. J. Mol. Sci. 2019, 20, 1031. https://doi.org/10.3390/ijms20051031
Fujita N, Kazama Y, Yamagishi N, Watanabe K, Ando S, Tsuji H, Kawano S, Yoshikawa N, Komatsu K. Development of the VIGS System in the Dioecious Plant Silene latifolia. International Journal of Molecular Sciences. 2019; 20(5):1031. https://doi.org/10.3390/ijms20051031
Chicago/Turabian StyleFujita, Naoko, Yusuke Kazama, Noriko Yamagishi, Kyoko Watanabe, Saki Ando, Hiroyuki Tsuji, Shigeyuki Kawano, Nobuyuki Yoshikawa, and Ken Komatsu. 2019. "Development of the VIGS System in the Dioecious Plant Silene latifolia" International Journal of Molecular Sciences 20, no. 5: 1031. https://doi.org/10.3390/ijms20051031
APA StyleFujita, N., Kazama, Y., Yamagishi, N., Watanabe, K., Ando, S., Tsuji, H., Kawano, S., Yoshikawa, N., & Komatsu, K. (2019). Development of the VIGS System in the Dioecious Plant Silene latifolia. International Journal of Molecular Sciences, 20(5), 1031. https://doi.org/10.3390/ijms20051031