DNA Damage Stress: Cui Prodest?
Abstract
:1. Introduction
2. Cell Cycle Surveillance System
3. After Event Cleaning Job: RELEASE of the DNA Damage Checkpoint
4. DNA Damage and the Balance between Survival and Death
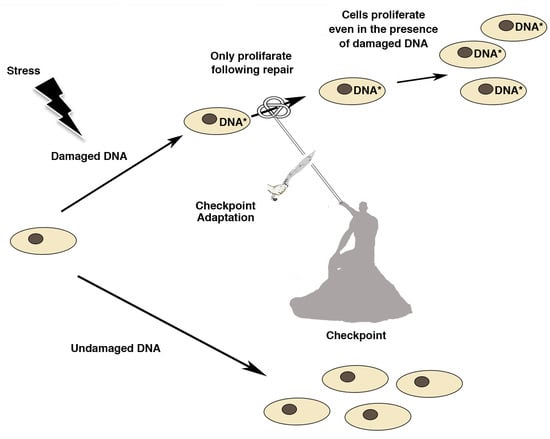
5. Molecular Mechanisms of Checkpoint Adaptation
6. Consequences of Checkpoint Adaptation
7. Future Directions
Funding
Conflicts of Interest
References
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, J.I.; Matsuoka, M. Oncogenic spiral by infectious pathogens: Cooperation of multiple factors in cancer development. Cancer Sci. 2018, 109, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Rue, P.; Martinez Arias, A. Cell dynamics and gene expression control in tissue homeostasis and development. Mol. Syst. Biol. 2015, 11, 792. [Google Scholar] [CrossRef] [PubMed]
- Chigira, M. Selfish cells in altruistic cell society—A theoretical oncology. Int. J. Oncol. 1993, 3, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Hersh, E.M.; Gutterman, J.U.; Mavligit, G.M. Cancer and host defense mechanisms. Pathobiol. Annu. 1975, 5, 133–167. [Google Scholar] [PubMed]
- Burgess, R.C.; Misteli, T. Not All DDRs Are Created Equal: Non-Canonical DNA Damage Responses. Cell 2015, 162, 944–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donzelli, M.; Draetta, G.F. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003, 4, 671–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, J.C.; Haber, J.E. Surviving the breakup: The DNA damage checkpoint. Annu. Rev. Genet. 2006, 40, 209–235. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007, 19, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, I.; Koepp, D.M. Recovery from the DNA Replication Checkpoint. Genes 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, I.A.; Krenning, L.; Bruinsma, W.; Medema, R.H. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 2015, 128, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Baldo, V.; Liang, J.; Wang, G.; Zhou, H. Preserving Yeast Genetic Heritage through DNA Damage Checkpoint Regulation and Telomere Maintenance. Biomolecules 2012, 2, 505–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syljuasen, R.G. Checkpoint adaptation in human cells. Oncogene 2007, 26, 5833–5839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair 2017, 56, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.W.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- Harper, J.W.; Elledge, S.J.; Keyomarsi, K.; Dynlacht, B.; Tsai, L.H.; Zhang, P.; Dobrowolski, S.; Bai, C.; Connell-Crowley, L.; Swindell, E.; et al. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 1995, 6, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Stracker, T.H.; Usui, T.; Petrini, J.H. Taking the time to make important decisions: The checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair 2009, 8, 1047–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niida, H.; Murata, K.; Shimada, M.; Ogawa, K.; Ohta, K.; Suzuki, K.; Fujigaki, H.; Khaw, A.K.; Banerjee, B.; Hande, M.P.; et al. Cooperative functions of Chk1 and Chk2 reduce tumour susceptibility in vivo. EMBO J. 2010, 29, 3558–3570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, F.; Clouaire, T.; Aguirrebengoa, M.; Legube, G.; Miller, K.M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 2017, 216, 1959–1974. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Vyas, A.; Kassab, M.A.; Singh, A.K.; Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 2017, 45, 8129–8141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macurek, L.; Lindqvist, A.; Lim, D.; Lampson, M.A.; Klompmaker, R.; Freire, R.; Clouin, C.; Taylor, S.S.; Yaffe, M.B.; Medema, R.H. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 2008, 455, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Boddy, M.N. Cooperativity of the SUMO and Ubiquitin Pathways in Genome Stability. Biomolecules 2016, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Piekna-Przybylska, D.; Bambara, R.A.; Balakrishnan, L. Acetylation regulates DNA repair mechanisms in human cells. Cell Cycle 2016, 15, 1506–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, H.X.; Poovey, C.E.; Privette, A.A.; Grant, G.D.; Chao, H.Y.; Cook, J.G.; Purvis, J.E. Orchestration of DNA Damage Checkpoint Dynamics across the Human Cell Cycle. Cell Syst. 2017, 5, 445–459.e5. [Google Scholar] [CrossRef] [PubMed]
- Giunta, S.; Belotserkovskaya, R.; Jackson, S.P. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 2010, 190, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, A.; de Bruijn, M.; Macurek, L.; Bras, A.; Mensinga, A.; Bruinsma, W.; Voets, O.; Kranenburg, O.; Medema, R.H. Wip1 confers G2 checkpoint recovery competence by counteracting p53-dependent transcriptional repression. EMBO J. 2009, 28, 3196–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pintard, L.; Archambault, V. A unified view of spatio-temporal control of mitotic entry: Polo kinase as the key. Open Biol. 2018, 8, 180114. [Google Scholar] [CrossRef] [PubMed]
- Lock, R.B.; Ross, W.E. Possible role for p34cdc2 kinase in etoposide-induced cell death of Chinese hamster ovary cells. Cancer Res. 1990, 50, 3767–3771. [Google Scholar] [PubMed]
- Smits, V.A.; Klompmaker, R.; Arnaud, L.; Rijksen, G.; Nigg, E.A.; Medema, R.H. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000, 2, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Smits, V.A.; Klompmaker, R.; Vallenius, T.; Rijksen, G.; Makela, T.P.; Medema, R.H. p21 inhibits Thr161 phosphorylation of Cdc2 to enforce the G2 DNA damage checkpoint. J. Biol. Chem. 2000, 275, 30638–30643. [Google Scholar] [CrossRef] [PubMed]
- Falck, J.; Lukas, C.; Protopopova, M.; Lukas, J.; Selivanova, G.; Bartek, J. Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene 2001, 20, 5503–5510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, B.; Gao, B.; Yu, J.; Yuan, J.; Lou, Z. Ataxia telangiectasia-mutated- and Rad3-related protein regulates the DNA damage-induced G2/M checkpoint through the Aurora A cofactor Bora protein. J. Biol. Chem. 2013, 288, 16139–16144. [Google Scholar] [CrossRef] [PubMed]
- Van Vugt, M.A.; Bras, A.; Medema, R.H. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell 2004, 15, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Carmena, M. Polo-like kinase-activating kinases: Aurora A, Aurora B and what else? Cell Cycle 2012, 11, 1490–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sur, S.; Agrawal, D.K. Phosphatases and kinases regulating CDC25 activity in the cell cycle: Clinical implications of CDC25 overexpression and potential treatment strategies. Mol. Cell. Biochem. 2016, 416, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Jullien, D.; Bugler, B.; Dozier, C.; Cazales, M.; Ducommun, B. Identification of N-terminally truncated stable nuclear isoforms of CDC25B that are specifically involved in G2/M checkpoint recovery. Cancer Res. 2011, 71, 1968–1977. [Google Scholar] [CrossRef] [PubMed]
- Van Vugt, M.A.; Gardino, A.K.; Linding, R.; Ostheimer, G.J.; Reinhardt, H.C.; Ong, S.E.; Tan, C.S.; Miao, H.; Keezer, S.M.; Li, J.; et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010, 8, e1000287. [Google Scholar] [CrossRef] [PubMed]
- Freire, R.; van Vugt, M.A.; Mamely, I.; Medema, R.H. Claspin: Timing the cell cycle arrest when the genome is damaged. Cell Cycle 2006, 5, 2831–2834. [Google Scholar] [CrossRef] [PubMed]
- Mamely, I.; van Vugt, M.A.; Smits, V.A.; Semple, J.I.; Lemmens, B.; Perrakis, A.; Medema, R.H.; Freire, R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 2006, 16, 1950–1955. [Google Scholar] [CrossRef] [PubMed]
- Peschiaroli, A.; Dorrello, N.V.; Guardavaccaro, D.; Venere, M.; Halazonetis, T.; Sherman, N.E.; Pagano, M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 2006, 23, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Borges, H.L.; Linden, R.; Wang, J.Y. DNA damage-induced cell death: Lessons from the central nervous system. Cell Res. 2008, 18, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Kracikova, M.; Akiri, G.; George, A.; Sachidanandam, R.; Aaronson, S.A. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 2013, 20, 576–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Ozaki, T.; Yamamoto, H.; Furuya, K.; Hosoda, M.; Hayashi, S.; Fukuzawa, M.; Nakagawara, A. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J. Biol. Chem. 2004, 279, 25549–25561. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Minsky, N.; Oren, M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell 2004, 16, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhao, W.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation is indispensable for p53 activation. Cell 2008, 133, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Gu, W. Dual Roles of MDM2 in the Regulation of p53: Ubiquitination Dependent and Ubiquitination Independent Mechanisms of MDM2 Repression of p53 Activity. Genes Cancer 2012, 3, 240–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Q.; Chen, J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle 2010, 9, 472–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puszynski, K.; Bertolusso, R.; Lipniacki, T. Crosstalk between p53 and nuclear factor-B systems: Pro- and anti-apoptotic functions of NF-B. IET Syst. Biol. 2009, 3, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Henrich, A.; Greiner, G.; Wolf, V.; Lovas, A.; Wieczorek, M.; Wagner, T.; Reichardt, S.; von Werder, A.; Schmid, R.M.; et al. Cross talk between stimulated NF-κB and the tumor suppressor p53. Oncogene 2010, 29, 2795–2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietsch, E.C.; Sykes, S.M.; McMahon, S.B.; Murphy, M.E. The p53 family and programmed cell death. Oncogene 2008, 27, 6507–6521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichwan, S.J.; Yamada, S.; Sumrejkanchanakij, P.; Ibrahim-Auerkari, E.; Eto, K.; Ikeda, M.A. Defect in serine 46 phosphorylation of p53 contributes to acquisition of p53 resistance in oral squamous cell carcinoma cells. Oncogene 2006, 25, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Mayo, L.D.; Seo, Y.R.; Jackson, M.W.; Smith, M.L.; Rivera Guzman, J.; Korgaonkar, C.K.; Donner, D.B. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J. Biol. Chem. 2005, 280, 25953–25959. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Arakawa, H.; Tanaka, T.; Matsuda, K.; Tanikawa, C.; Mori, T.; Nishimori, H.; Tamai, K.; Tokino, T.; Nakamura, Y.; et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000, 102, 849–862. [Google Scholar] [CrossRef]
- Bulavin, D.V.; Saito, S.; Hollander, M.C.; Sakaguchi, K.; Anderson, C.W.; Appella, E.; Fornace, A.J., Jr. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999, 18, 6845–6854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, T.G.; Moller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Droge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Taira, N.; Nihira, K.; Yamaguchi, T.; Miki, Y.; Yoshida, K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol. Cell 2007, 25, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Toczyski, D.P.; Galgoczy, D.J.; Hartwell, L.H. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 1997, 90, 1097–1106. [Google Scholar] [CrossRef]
- Yoshida, K.; Liu, H.; Miki, Y. Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J. Biol. Chem. 2006, 281, 5734–5740. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.B.; Schumacher, B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulovich, A.G.; Margulies, R.U.; Garvik, B.M.; Hartwell, L.H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 1997, 145, 45–62. [Google Scholar] [PubMed]
- Paulovich, A.G.; Toczyski, D.P.; Hartwell, L.H. When checkpoints fail. Cell 1997, 88, 315–321. [Google Scholar] [CrossRef]
- Sandell, L.L.; Zakian, V.A. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell 1993, 75, 729–739. [Google Scholar] [CrossRef]
- Pardo, B.; Crabbe, L.; Pasero, P. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Leroy, C.; Lee, S.E.; Vaze, M.B.; Ochsenbein, F.; Guerois, R.; Haber, J.E.; Marsolier-Kergoat, M.C. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 2003, 11, 827–835. [Google Scholar] [CrossRef]
- Pellicioli, A.; Lee, S.E.; Lucca, C.; Foiani, M.; Haber, J.E. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 2001, 7, 293–300. [Google Scholar] [CrossRef]
- Vidanes, G.M.; Sweeney, F.D.; Galicia, S.; Cheung, S.; Doyle, J.P.; Durocher, D.; Toczyski, D.P. CDC5 inhibits the hyperphosphorylation of the checkpoint kinase Rad53, leading to checkpoint adaptation. PLoS Biol. 2010, 8, e1000286. [Google Scholar] [CrossRef] [PubMed]
- Guillemain, G.; Ma, E.; Mauger, S.; Miron, S.; Thai, R.; Guerois, R.; Ochsenbein, F.; Marsolier-Kergoat, M.C. Mechanisms of checkpoint kinase Rad53 inactivation after a double-strand break in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 3378–3389. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 2004, 117, 575–588. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Cimprich, K.A. Checkpoint adaptation; molecular mechanisms uncovered. Cell 2004, 117, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Bekker-Jensen, S.; Bartek, J.; Lukas, J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 2006, 23, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Bassermann, F.; Frescas, D.; Guardavaccaro, D.; Busino, L.; Peschiaroli, A.; Pagano, M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 2008, 134, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Esposito, A.; De, S.; Ber, S.; Collin, P.; Surana, U.; Venkitaraman, A.R. Homeostatic control of polo-like kinase-1 engenders non-genetic heterogeneity in G2 checkpoint fidelity and timing. Nat. Commun. 2014, 5, 4048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, H.; Benada, J.; Mullers, E.; Akopyan, K.; Burdova, K.; Koolmeister, T.; Helleday, T.; Medema, R.H.; Macurek, L.; Lindqvist, A. ATM/Wip1 activities at chromatin control Plk1 re-activation to determine G2 checkpoint duration. EMBO J. 2017, 36, 2161–2176. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. DNA repair: Damage alert. Nature 2003, 421, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Van Vugt, M.A.; Bras, A.; Medema, R.H. Restarting the cell cycle when the checkpoint comes to a halt. Cancer Res. 2005, 65, 7037–7040. [Google Scholar] [CrossRef] [PubMed]
- Bender, K.; Vydzhak, O.; Klermund, J.; Busch, A.; Grimm, S.; Luke, B. Checkpoint adaptation in repair-deficient cells drives aneuploidy and resistance to genotoxic agents. BioRxiv 2018. [Google Scholar] [CrossRef]
- Lewis, C.W.; Golsteyn, R.M. Cancer cells that survive checkpoint adaptation contain micronuclei that harbor damaged DNA. Cell Cycle 2016, 15, 3131–3145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubara, P.M.; Kerneis-Golsteyn, S.; Studeny, A.; Lanser, B.B.; Meijer, L.; Golsteyn, R.M. Human cells enter mitosis with damaged DNA after treatment with pharmacological concentrations of genotoxic agents. Biochem. J. 2012, 446, 373–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swift, L.H.; Golsteyn, R.M. Cytotoxic amounts of cisplatin induce either checkpoint adaptation or apoptosis in a concentration-dependent manner in cancer cells. Biol. Cell 2016, 108, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Galgoczy, D.J.; Toczyski, D.P. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 2001, 21, 1710–1718. [Google Scholar] [CrossRef] [PubMed]
- Syljuasen, R.G.; Jensen, S.; Bartek, J.; Lukas, J. Adaptation to the ionizing radiation-induced G2 checkpoint occurs in human cells and depends on checkpoint kinase 1 and Polo-like kinase 1 kinases. Cancer Res. 2006, 66, 10253–10257. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Xing, L.; Shen, H.; Lian, W.; Yi, L.; Zhang, D.; Yang, H.; Liu, J.; Zhang, X. Sterigmatocystin-induced checkpoint adaptation depends on Chk1 in immortalized human gastric epithelial cells in vitro. Arch. Toxicol. 2017, 91, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.A.; Cleveland, D.W. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell 2005, 8, 7–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swift, L.H.; Golsteyn, R.M. Genotoxic anti-cancer agents and their relationship to DNA damage, mitosis, and checkpoint adaptation in proliferating cancer cells. Int. J. Mol. Sci. 2014, 15, 3403–3431. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Berdis, A. Combating resistance to DNA damaging agents. Oncoscience 2018, 5, 134–136. [Google Scholar] [PubMed]
- Sprouffske, K.; Merlo, L.M.; Gerrish, P.J.; Maley, C.C.; Sniegowski, P.D. Cancer in light of experimental evolution. Curr. Biol. 2012, 22, R762–R771. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.B.; Johnson, L.J.; Jackson, R.W.; Brockhurst, M.A.; Dash, P.R. First steps in experimental cancer evolution. Evol. Appl. 2013, 6, 535–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, T.B.; Wass, A.V.; Johnson, L.J.; Dash, P. Resource competition promotes tumour expansion in experimentally evolved cancer. BMC Evol. Biol. 2017, 17, 268. [Google Scholar] [CrossRef] [PubMed]
- Malaquin, N.; Carrier-Leclerc, A.; Dessureault, M.; Rodier, F. DDR-mediated crosstalk between DNA-damaged cells and their microenvironment. Front. Genet. 2015, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Cialfi, S.; Le Pera, L.; De Blasio, C.; Mariano, G.; Palermo, R.; Zonfrilli, A.; Uccelletti, D.; Palleschi, C.; Biolcati, G.; Barbieri, L.; et al. The loss of ATP2C1 impairs the DNA damage response and induces altered skin homeostasis: Consequences for epidermal biology in Hailey-Hailey disease. Sci. Rep. 2016, 6, 31567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horwitz, E.; Krogvold, L.; Zhitomirsky, S.; Swisa, A.; Fischman, M.; Lax, T.; Dahan, T.; Hurvitz, N.; Weinberg-Corem, N.; Klochendler, A.; et al. β-Cell DNA Damage Response Promotes Islet Inflammation in Type 1 Diabetes. Diabetes 2018, 67, 2305–2318. [Google Scholar] [CrossRef] [PubMed]
- Milanese, C.; Cerri, S.; Ulusoy, A.; Gornati, S.V.; Plat, A.; Gabriels, S.; Blandini, F.; Di Monte, D.A.; Hoeijmakers, J.H.; Mastroberardino, P.G. Activation of the DNA damage response in vivo in synucleinopathy models of Parkinson’s disease. Cell Death Dis. 2018, 9, 818. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.; Murphy, M.; Garwood, C.J.; Jennings, L.; Heath, P.R.; Chambers, A.; Matthews, F.E.; Brayne, C.; Ince, P.G.; Wharton, S.B.; et al. Metallothionein-I/II expression associates with the astrocyte DNA damage response and not Alzheimer-type pathology in the aging brain. Glia 2018, 66, 2316–2323. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, S.; Oaks, J.; Ren, J.; Li, L.; Wu, X. The concerted roles of FANCM and Rad52 in the protection of common fragile sites. Nat. Commun. 2018, 9, 2791. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, N.; Franchitto, M.; Zonfrilli, A.; Cialfi, S.; Palermo, R.; Talora, C. DNA Damage Stress: Cui Prodest? Int. J. Mol. Sci. 2019, 20, 1073. https://doi.org/10.3390/ijms20051073
Verma N, Franchitto M, Zonfrilli A, Cialfi S, Palermo R, Talora C. DNA Damage Stress: Cui Prodest? International Journal of Molecular Sciences. 2019; 20(5):1073. https://doi.org/10.3390/ijms20051073
Chicago/Turabian StyleVerma, Nagendra, Matteo Franchitto, Azzurra Zonfrilli, Samantha Cialfi, Rocco Palermo, and Claudio Talora. 2019. "DNA Damage Stress: Cui Prodest?" International Journal of Molecular Sciences 20, no. 5: 1073. https://doi.org/10.3390/ijms20051073