On a Cold Night: Transcriptomics of Grapevine Flower Unveils Signal Transduction and Impacted Metabolism
Abstract
:1. Introduction
2. Results and Discussion
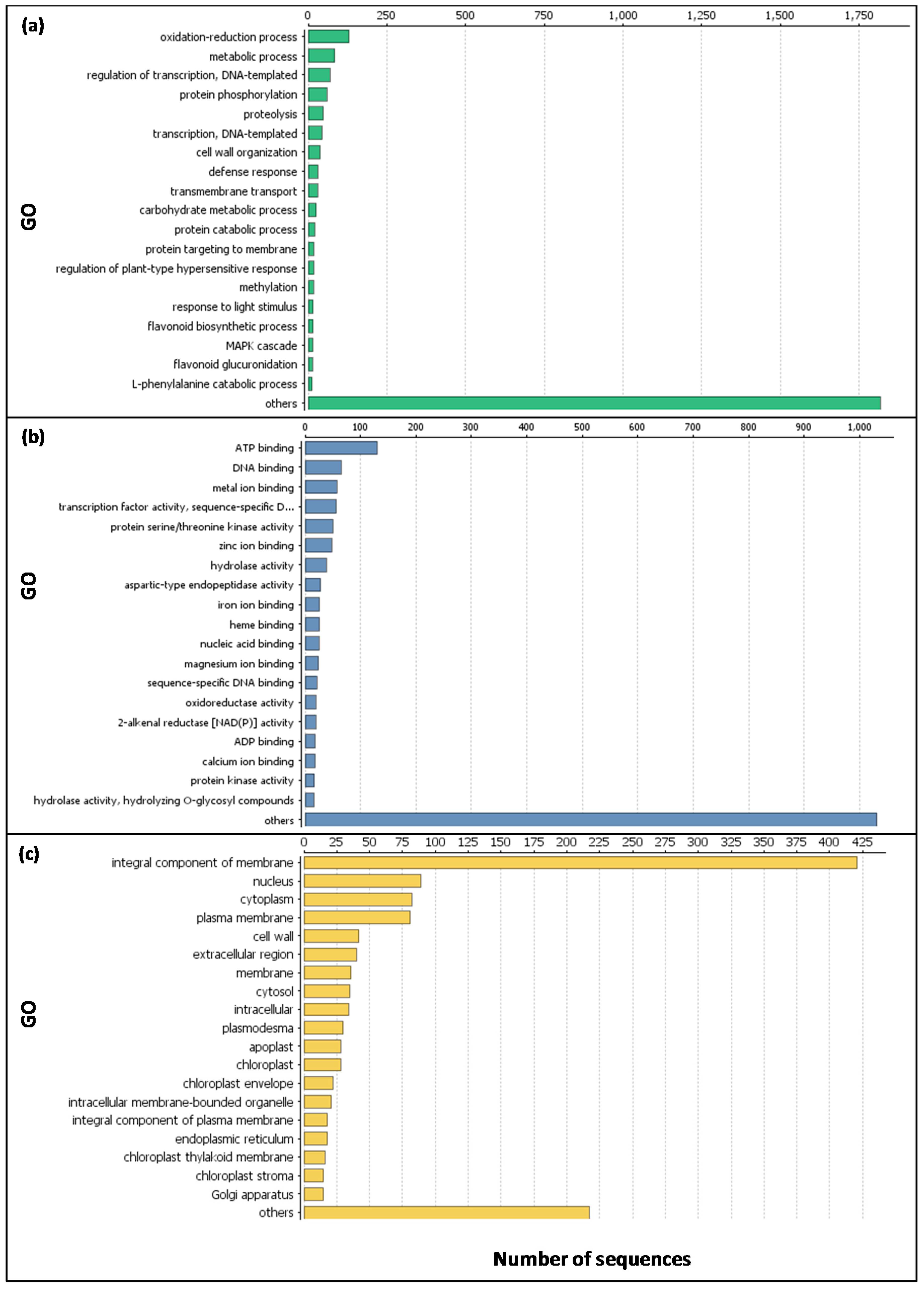
2.1. Expression Analysis and Validation of the Data Set
2.2. Functional Analyses of the Transcriptome Changes in Response to a Cold Night
2.3. Cold Stress Perception by the Grapevine Flower and Signal Transduction
2.4. Calcium-CaM and some Kinases Encoding Genes are Overexpressed by Cold Stress
2.5. Transcription Factors
2.5.1. Genes Encoding CBF Transcription Factors are Induced by Cold Stress
2.5.2. Cold Stress Affects the Expression of Genes Encoding Transcription Factors Involved in the Circadian Clock
2.5.3. Heat-Shock Factors
2.5.4. Cold Impacts the Expression of EDS1, SGT1, SAG101, TIR-NB-LRR, PHI-1, Genes Reported to be Involved in Global Stress Responses
2.6. Cold Modulates the Expression of Genes Involved in Global Stress Responses
2.6.1. Cold Night Induces an Over-Expression of Genes Encoding Chaperone Proteins
2.6.2. Transcripts Involved in Detoxification Pathways are Induced by Cold Stress in Grapevine Flowers
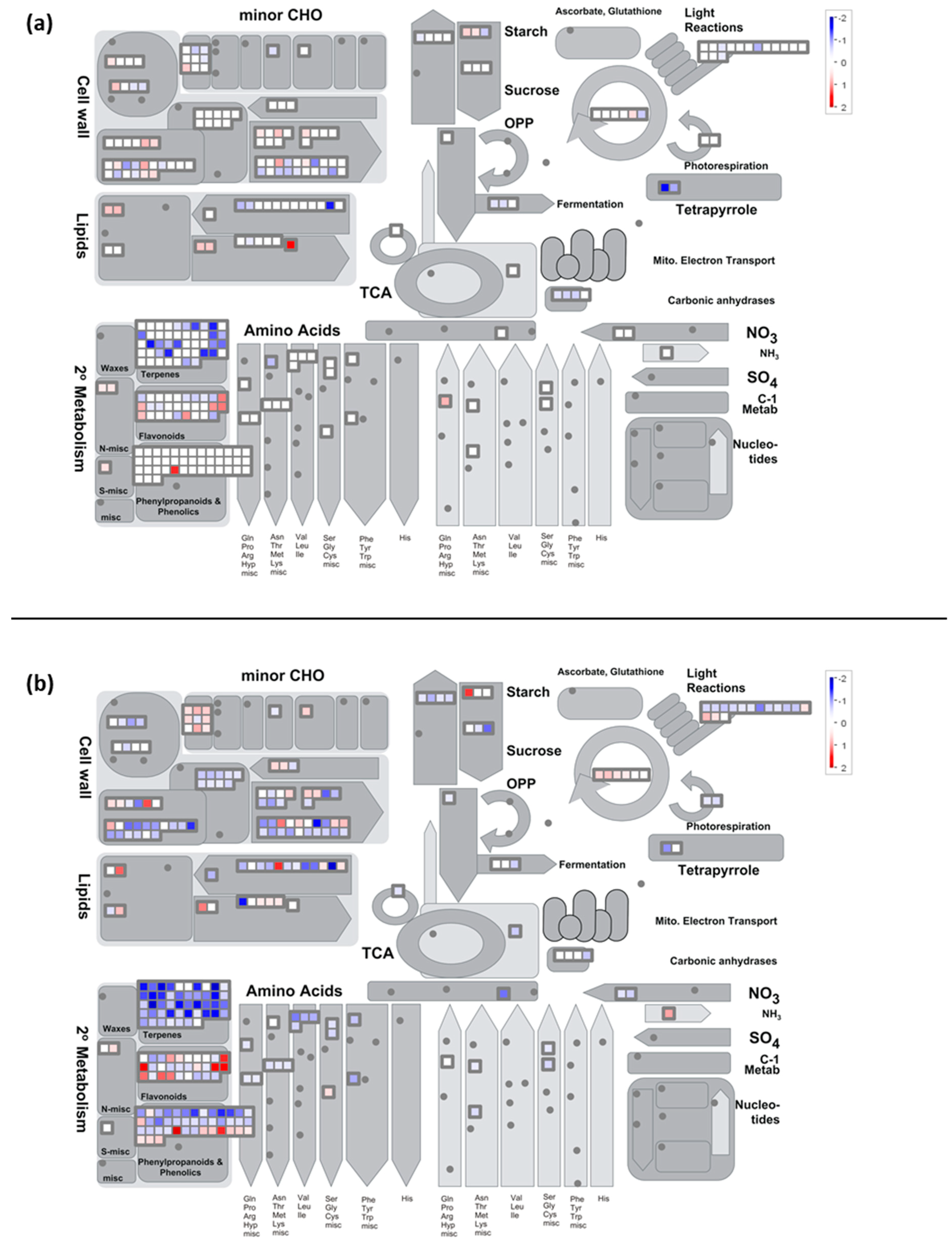
2.7. Metabolisms Impaired by the Cold Night
2.7.1. Carbon Metabolism
- Cold stress impacts the expression of genes encoding proteins of the photosystem antenna complexes.
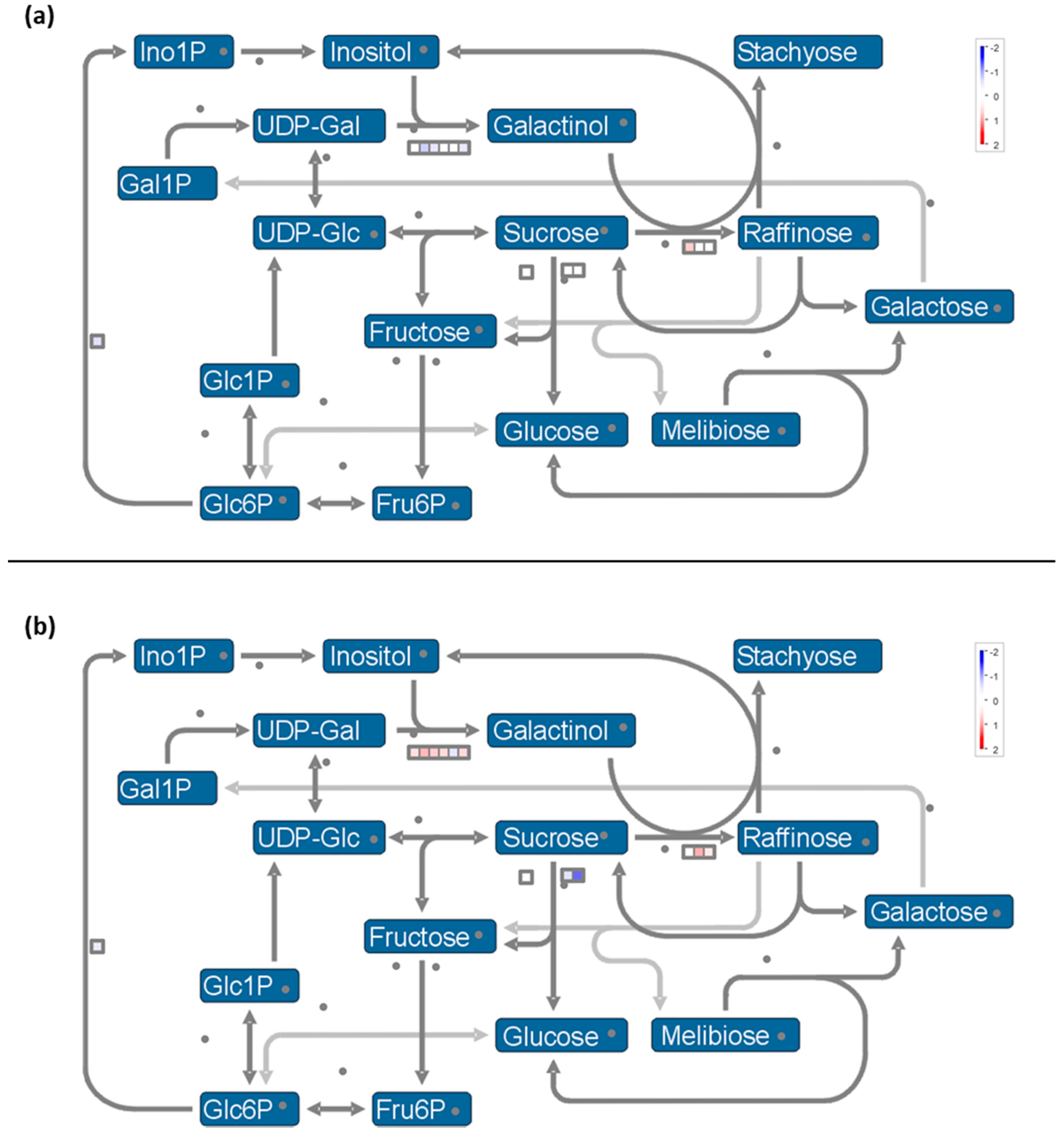
- Expression of genes involved in synthesis of osmoprotectant sugars are differently impacted during stress and recovery
2.7.2. Cold Modulates the Expression of Genes Encoding Components Involved in Cell Wall Degradation and Synthesis
2.7.3. Secondary Metabolism
- Cold down-regulates the expression of genes related to the flavonoid pathway
- The expression of valencene synthase encoding gene is down-regulated by the cold night
3. Materials and Methods
3.1. Plant Materials and Cold Treatment
3.2. RNA Extraction
3.3. Microarray Analysis
3.4. Statistical Analysis of Microarray Data
3.5. Data Deposition
3.6. Transcriptome Analysis
3.7. Functional Analyses
3.8. Real-Time PCR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LD4CL | 4-coumarate:coA ligase |
| ABA | Abscisic acid |
| AGPase | AdenosineDiPhosphate-glucose pyrophosphorylase |
| AP2/EREBP | Ethylene-Responsive Element Binding Protein |
| ARR | Arabidospsis thaliana response regulator |
| ATPA2 | ATP synthase subunit alpha 2 |
| bHLH | Basic Helix-Loop-Helix |
| BIN | Mapman classes |
| CAD/SAD | Cinnamyl/Sinapyl Alcohol Dehydrogenase |
| CaM | Calmodulin |
| CAMTA3 | Calmodulin binding transcription activator 3 |
| CBF | C-repeat/DRE-Binding Factor |
| CBF/DREB | C-repeat binding factor/ Dehydration-responsive element binding |
| CCoAOMT | Caffeoyl-CoA O-methyltransferase |
| CDPK | (Ca2+)-dependent protein kinases |
| CHI4C | Class IV chitinase |
| CHO | Carbohydrates |
| CO-like | Constans-like zinc finger family |
| COR | Cold-inducible |
| CRT | Calreticulin |
| Cyt b559 | Cytochrome b559 |
| DEG | Differentially Expressed Genes |
| EDS1 | Enhanced Disease Susceptibility 1 |
| ERD | Early Response to Dehydration |
| ERF/AP2 | Ethylene Responsive Factor/APETALA2 |
| ERFs | Ethylene Response Factors |
| GIN1 | Vacuolar invertases 1 |
| GO | Gene Ontology |
| GOLS | Galactinol synthase |
| GST | Glutathione S-Transferase |
| HSP | Heat Shock Proteins |
| HVA22 | ABA- and stress-inducible gene first isolated from barley |
| ICE | Inducer of CBF Expression |
| LAC4 | Laccase 4 |
| LEA | Late-Embryogenesis Abundant |
| LHCB | Light-Harvesting Chlorophyll a/b Binding |
| LHY | Late elongated hypocotyl |
| LRK10 | Like locus receptor-like protein kinase 10 |
| MAPK | Mitogen-Activated Protein Kinases |
| MAPKK2 | Mitogen-Activated Protein Kinase Kinase 2 |
| MLO | Mildew resistance Locus O |
| MPKKK5 | MAP Kinase Kinase Kinase 5 |
| MYB | Myeloblastosis |
| NAC | No Apical Meristem |
| Ndh | NADH dehydrogenase |
| PAD4 | PhytoAlexin Deficient 4 |
| PAL | Phenylalanine Ammonia Lyase |
| PG | PolyGalacturonase |
| PR | Pathogen-Related |
| PRR | Pseudo Response Regulator |
| PS | Post-Stress |
| PSI | PhotoSystem I |
| PSII | PhotoSystem II |
| RFOs | Raffinose Family Oligosaccharides |
| ROS | Reactive Oxygen Species |
| RPK | Receptor serine/threonine kinase and receptor-like Protein Kinase |
| RS | Raffinose Synthase |
| RT-qPCR | Quantitative real-time RT-PCR |
| RVE1 | Protein reveille 1 |
| SA | Salicylic Acid |
| SAG101 | Senescence Associated Gene 101 |
| SCUTL2 | Thaumatin-like protein |
| SGT1 | Suppressor of G2 Allele SKP1 |
| STS | Stilbene Synthase |
| TF | Transcription factors |
| TIR-NB-LRR | Toll-interleukin-1 receptor-nucleotide binding-leucine rich repeat linear dichroism |
References
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef]
- Lv, D.K.; Bai, X.; Li, Y.; Ding, X.D.; Ge, Y.; Cai, H.; Ji, W.; Wu, N.; Zhu, Y.M. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 2010, 459, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Gill, S.S.; Tuteja, R. Plant Responses to Abiotic Stresses: Shedding Light on Salt, Drought, Cold and Heavy Metal Stress. In Omics and Plant Abiotic Stress Tolerance; Bentham Science Publishers: Sharjah, UAE, 2011; pp. 39–64. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Steponkus, P.L. Role of the Plasma Membrane in Freezing Injury and Cold Acclimation. Annu. Rev. Plant Biol. 1984, 35, 543–584. [Google Scholar] [CrossRef]
- Su, F.; Gilard, F.; Guerard, F.; Citerne, S.; Clément, C.; Vaillant-Gaveau, N.; Dhondt-Cordelier, S. Spatio-temporal Responses of Arabidopsis Leaves in Photosynthetic Performance and Metabolite Contents to Burkholderia phytofirmans PsJN. Front. Plant Sci. 2016, 7, 403. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Zhao, Q.Y.; Ma, C.L.; Zhang, Z.H.; Cao, H.L.; Kong, Y.M.; Yue, C.; Hao, X.Y.; Chen, L.; Ma, J.Q.; et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genom. 2013, 14, 415. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Sung, D.Y.; Zhao, W.; Popp, M.; Porat, R.; Guy, C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007, 50, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.V.; Li, Q.B.; Haskell, D.W.; Guy, C.L. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994, 104, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P.; Sacco, M.; Cherutti, J.F.; Hill, S. Cold-Induced Accumulation of hsp90 Transcripts in Brassica napus. Plant Physiol. 1995, 107, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar] [PubMed]
- Yang, T.; Chaudhuri, S.; Yang, L.; Du, L.; Poovaiah, B.W. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 2010, 285, 7119–7126. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Ruelland, E.; Vaultier, M.-N.; Zachowski, A.; Hurry, V. Cold Signalling and Cold Acclimation in Plants. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2009; Volume 49, pp. 35–150. [Google Scholar]
- Szabados, L.; Savoure, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Matsuda, O.; Sakamoto, H.; Hashimoto, T.; Iba, K. A temperature-sensitive mechanism that regulates post-translational stability of a plastidial omega-3 fatty acid desaturase (FAD8) in Arabidopsis leaf tissues. J. Biol. Chem. 2005, 280, 3597–3604. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Fuller, M.P.; Telli, G. An investigation of the frost hardiness of grapevine (Vitis vinifera) during bud break. Ann. Appl. Biol. 1999, 135, 589–595. [Google Scholar] [CrossRef]
- Lebon, G.; Duchêne, E.; Brun, O.; Magné, C.; Clément, C. Flower abscission and inflorescence carbohydrates in sensitive and non-sensitive cultivars of grapevine. Sex. Plant Reprod. 2004, 17, 71–79. [Google Scholar] [CrossRef]
- Allen, D.J.; Ort, D.R. Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci. 2001, 6, 36–42. [Google Scholar] [CrossRef]
- Bertamini, M.; Muthuchelian, K.; Rubinigg, M.; Zorer, R.; Nedunchezhian, N. Photoinhibition of photosynthesis in leaves of grapevine (Vitis vinifera L. cv. Riesling). Effect of chilling nights. Photosynthetica 2005, 43, 551–557. [Google Scholar] [CrossRef]
- Hendrickson, L.; Ball, M.C.; Wood, J.T.; Chow, W.S.; Furbank, R.T. Low temperature effects on photosynthesis and growth of grapevine. Plant Cell Environ. 2004, 27, 795–809. [Google Scholar] [CrossRef]
- Sawicki, M.; Jeanson, E.; Celiz, V.; Clément, C.; Jacquard, C.; Vaillant-Gaveau, N. Adaptation of grapevine flowers to cold involves different mechanisms depending on stress intensity. PLoS ONE 2012, 7, e46976. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, O.; Vandesteene, L.; Feil, R.; Baillieul, F.; Lunn, J.E.; Clément, C. Trehalose metabolism is activated upon chilling in grapevine and might participate in Burkholderia phytofirmans induced chilling tolerance. Planta 2012, 236, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, M.; Ait Barka, E.; Clément, C.; Gilard, F.; Tcherkez, G.; Baillieul, F.; Vaillant-Gaveau, N.; Jacquard, C. Cold-night responses in grapevine inflorescences. Plant Sci. 2015, 239, 115–127. [Google Scholar] [CrossRef]
- Aschan, G.; Pfanz, H. Non-foliar photosynthesis-a strategy of additional carbon acquisition. Flora 2003, 198, 81–97. [Google Scholar] [CrossRef]
- Sawicki, M.; Courteaux, B.; Rabenoelina, F.; Baillieul, F.; Clément, C.; Ait Barka, E.; Jacquard, C.; Vaillant-Gaveau, N. Leaf vs. inflorescence: Differences in photosynthetic activity of grapevine. Photosynthetica 2017, 55, 58–68. [Google Scholar] [CrossRef]
- Vaillant-Gaveau, N.; Maillard, P.; Wojnarowiez, G.; Gross, P.; Clément, C.; Fontaine, F. Inflorescence of grapevine (Vitis vinifera L.): A high ability to distribute its own assimilates. J. Exp. Bot. 2011, 62, 4183–4190. [Google Scholar] [CrossRef]
- Tattersall, E.A.; Grimplet, J.; DeLuc, L.; Wheatley, M.D.; Vincent, D.; Osborne, C.; Ergul, A.; Lomen, E.; Blank, R.R.; Schlauch, K.A.; et al. Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Funct. Integr. Genom. 2007, 7, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Zhu, W.; Wang, L.; Xiang, Y.; Fang, L.; Li, J.; Sun, X.; Wang, N.; Londo, J.P.; Li, S. Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress. PLoS ONE 2013, 8, e58740. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Zhang, Y.L.; Shao, H.; Lu, J. Differential Physio-Biochemical Responses to Cold Stress of Cold-Tolerant and Non-Tolerant Grapes (Vitis L.) from China. J. Agron. Crop Sci. 2010, 196, 212–219. [Google Scholar] [CrossRef]
- Rowland, L.J.; Alkharouf, N.; Darwish, O.; Ogden, E.L.; Polashock, J.J.; Bassil, N.V.; Main, D. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Arora, R. Mechanism of freeze-thaw injury and recovery: A cool retrospective and warming up to new ideas. Plant Sci. 2018, 270, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.H.; Hu, X.; Tang, W.; Zheng, X.; Kim, Y.S.; Lee, B.H.; Zhu, J.K. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 2006, 26, 9533–9543. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Lee, H.; Xiong, L.; Jagendorf, A.; Stevenson, B.; Zhu, J.K. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 2002, 99, 11507–11512. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002, 129, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Arisz, S.A.; van Wijk, R.; Roels, W.; Zhu, J.K.; Haring, M.A.; Munnik, T. Rapid phosphatidic acid accumulation in response to low temperature stress in Arabidopsis is generated through diacylglycerol kinase. Front. Plant Sci. 2013, 4, 1. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Campoli, C.; Matus-Cadiz, M.A.; Pozniak, C.J.; Cattivelli, L.; Fowler, D.B. Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol. Genet. Genom. 2009, 282, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Bordiec, S.; Fernandez, O.; Paquis, S.; Dhondt-Cordelier, S.; Baillieul, F.; Clément, C.; Barka, E.A. Burkholderia phytofirmans PsJN primes Vitis vinifera L. and confers a better tolerance to low nonfreezing temperatures. Mol. Plant Microbe Interact 2012, 25, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.; Kaplan, F.; Kopka, J.; Selbig, J.; Hincha, D.K. Metabolomics of temperature stress. Physiol. Plant. 2008, 132, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 2006, 126, 62–71. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.K. Abiotic stress signal transduction in plants: Molecular and genetic perspectives. Physiol. Plant. 2001, 112, 152–166. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Schumaker, K.; Zhu, J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004, 55, 225–236. [Google Scholar] [CrossRef]
- Cook, D.; Fowler, S.; Fiehn, O.; Thomashow, M.F. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 15243–15248. [Google Scholar] [CrossRef]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Van Der Luit, A.H.; Olivari, C.; Haley, A.; Knight, M.R.; Trewavas, A.J. Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol. 1999, 121, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yang, G.; Khan, M.; Onodera, H.; Toki, S.; Yamaguchi, M. Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genom. 2007, 277, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Onodera, H.; Ugaki, M.; Tanaka, H.; Komatsu, S. Characterization of calreticulin as a phosphoprotein interacting with cold-induced protein kinase in rice. Biol. Pharm. Bull. 2003, 26, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Jon, J.H.; Kwak, J.M.; Nam, H.G. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997, 113, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.R.; Walker, J.C. Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 2003, 6, 339–342. [Google Scholar] [CrossRef]
- Hardie, D.G. Plant Protein Serine/Threonine Kinases: Classification and Functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 97–131. [Google Scholar] [CrossRef]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 2014, 166, 2100–2114. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Grill, E.; Meskiene, I.; Schweighofer, A. Type 2C protein phosphatases in plants. FEBS J. 2013, 280, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Carlow, C. Analysis of the Vitis C-Repeat Binding Factor (CBF) Genes and Their Potential Roles in Both the CBF and Stomatal Development Path Ways. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2016. [Google Scholar]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Tattersall, E.A.R.; Siddiqua, M.K.; Cramer, G.R.; Nassuth, A. CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 2008, 31, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Zhang, M.; Yu, Z.; Ren, J.; Qin, Y.; Wang, B.; Xiao, L.; Zhang, Z.; Tao, J. Isolation and expression analysis of CBF4 from Vitis amurensis associated with stress. Agric. Sci. 2013, 4, 224–229. [Google Scholar]
- Takuhara, Y.; Kobayashi, M.; Suzuki, S. Low-temperature-induced transcription factors in grapevine enhance cold tolerance in transgenic Arabidopsis plants. J. Plant Physiol. 2011, 168, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Takao, S.; Kudo, T.; Kiba, T.; Wang, Y.; Kinoshita, T.; Sakakibara, H. Improvement of Arabidopsis Biomass and Cold, Drought and Salinity Stress Tolerance by Modified Circadian Clock-Associated PSEUDO-RESPONSE REGULATORs. Plant Cell Physiol. 2016, 57, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Kinmonth-Schultz, H.A.; Tong, X.; Lee, J.; Song, Y.H.; Ito, S.; Kim, S.H.; Imaizumi, T. Cool night-time temperatures induce the expression of CONSTANS and FLOWERING LOCUS T to regulate flowering in Arabidopsis. New Phytol. 2016, 211, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Meissner, M.; Orsini, E.; Ruschhaupt, M.; Melchinger, A.E.; Hincha, D.K.; Heyer, A.G. Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation. Plant Cell Environ. 2013, 36, 1256–1267. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005, 1, e26. [Google Scholar] [CrossRef]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef]
- Venugopal, S.C.; Jeong, R.D.; Mandal, M.K.; Zhu, S.; Chandra-Shekara, A.C.; Xia, Y.; Hersh, M.; Stromberg, A.J.; Navarre, D.; Kachroo, A.; et al. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009, 5, e1000545. [Google Scholar] [CrossRef]
- Wiermer, M.; Feys, B.J.; Parker, J.E. Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 2005, 8, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Ochsenbein, C.; Przybyla, D.; Danon, A.; Landgraf, F.; Gobel, C.; Imboden, A.; Feussner, I.; Apel, K. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J. 2006, 47, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, A.; Thamilarasan, S.K.; Park, J.I.; Jung, M.Y.; Nou, I.S. Characterization and abiotic stress-responsive expression analysis of SGT1 genes in Brassica oleracea. Genome 2016, 59, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Rensink, W.; Hart, A.; Liu, J.; Ouyang, S.; Zismann, V.; Buell, C.R. Analyzing the potato abiotic stress transcriptome using expressed sequence tags. Genome 2005, 48, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.H.; Sheen, J. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Renaut, J.; Hausman, J.-F.; Wisniewski, M.E. Proteomics and low-temperature studies: Bridging the gap between gene expression and metabolism. Physiol. Plant. 2006, 126, 97–109. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteom. 2008, 71, 391–411. [Google Scholar] [CrossRef]
- Griffith, M.; Yaish, M.W. Antifreeze proteins in overwintering plants: A tale of two activities. Trends Plant Sci. 2004, 9, 399–405. [Google Scholar] [CrossRef]
- Fernandes, H.; Michalska, K.; Sikorski, M.; Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Liu, G.; Li, H. Cloning and Characterization of a Pathogenesis-Related Gene (ThPR10) from Tamarix hispida. Acta Biol. Crac. Ser. Bot. 2010, 52, 17–25. [Google Scholar] [CrossRef]
- Lee, O.R.; Pulla, R.K.; Kim, Y.J.; Balusamy, S.R.; Yang, D.C. Expression and stress tolerance of PR10 genes from Panax ginseng C. A. Meyer. Mol. Biol. Rep. 2012, 39, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Kalmar, E.; Torok, Z.; Tompa, P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008, 147, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Puhakainen, T.; Hess, M.W.; Mäkelä, P.; Svensson, J.; Heino, P.; Palva, E.T. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 2004, 54, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, M.; Zhu, Z.; Li, S.; Xu, Y.; Zhang, C.; Singer, S.D.; Wang, Y. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Allagulova, C.R.; Gimalov, F.R.; Shakirova, F.M.; Vakhitov, V.A. The plant dehydrins: Structure and putative functions. Biochemistry 2003, 68, 945–951. [Google Scholar] [PubMed]
- Renaut, J.; Lutts, S.; Hoffmann, L.; Hausman, J.F. Responses of poplar to chilling temperatures: Proteomic and physiological aspects. Plant Biol. 2004, 6, 81–90. [Google Scholar] [PubMed]
- Shen, Q.; Chen, C.N.; Brands, A.; Pan, S.M.; Ho, T.H. The stress- and abscisic acid-induced barley gene HVA22: Developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 2001, 45, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.-L. Physiological functional analysis of a stress-induced protein, HVA22, in Escherichia coli. Access Int. J. 2013, 1, 14–23. [Google Scholar]
- Kim, S.A.; Ahn, S.Y.; Yun, H.K. Transcriptome analysis of grapevine shoots exposed to chilling temperature for four weeks. Hortic. Environ. Biotechnol. 2016, 57, 161–172. [Google Scholar] [CrossRef]
- Østergaard, L.; Teilum, K.; Mirza, O.; Mattsson, O.; Petersen, M.; Welinder, K.G.; Mundy, J.; Gajhede, M.; Henriksen, A. Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 2000, 44, 231–243. [Google Scholar] [CrossRef]
- Jansson, S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999, 4, 236–240. [Google Scholar] [CrossRef]
- Wong, C.E.; Li, Y.; Labbe, A.; Guevara, D.; Nuin, P.; Whitty, B.; Diaz, C.; Golding, G.B.; Gray, G.R.; Weretilnyk, E.A.; et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006, 140, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J.; Walters, R.G.; Horton, P.; Jansson, S. Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. Plant Cell 2001, 13, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Heddad, M.; Adamska, I. Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol. 2003, 132, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Ganeteg, U.; Kulheim, C.; Andersson, J.; Jansson, S. Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 2004, 134, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Pruneda-Paz, J.L.; Kay, S.A. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010, 15, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Öquist, G.; Chow, W.S.; Anderson, J.M. Photoinhibition of photosynthesis represents a mechanism for the long-term regulation of photosystem II. Planta 1992, 186, 450–460. [Google Scholar] [CrossRef]
- Huner, N.P.A.; Öquist, G.; Sarhan, F. Energy balance and acclimation to light and cold. Trends Plant Sci. 1998, 3, 224–230. [Google Scholar] [CrossRef]
- Miyake, C.; Schreiber, U.; Asada, K. Ferredoxin-Dependent and Antimycin A-Sensitive Reduction of Cytochrome b-559 by Far-Red Light in Maize Thylakoids; Participation of a Menadiol-Reducible Cytochrome b-559 in Cyclic Electron Flow. Plant Cell Physiol. 1995, 36, 743–748. [Google Scholar]
- Miyake, C.; Yonekura, K.; Kobayashi, Y.; Yokota, A. Cyclic electron flow within PSII functions in intact chloroplasts from spinach leaves. Plant Cell Physiol. 2002, 43, 951–957. [Google Scholar] [CrossRef]
- Joët, T.; Cournac, L.; Horvath, E.M.; Medgyesy, P.; Peltier, G. Increased sensitivity of photosynthesis to antimycin A induced by inactivation of the chloroplast ndhB gene. Evidence for a participation of the NADH-dehydrogenase complex to cyclic electron flow around photosystem I. Plant Physiol. 2001, 125, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.-I.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004, 429, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Yang, S.J.; Zhang, S.B.; Zhang, J.L.; Cao, K.F. Cyclic electron flow plays an important role in photoprotection for the resurrection plant Paraboea rufescens under drought stress. Planta 2012, 235, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, S.B.; Cao, K.F. Cyclic electron flow plays an important role in photoprotection of tropical trees illuminated at temporal chilling temperature. Plant Cell Physiol. 2011, 52, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Pillet, J.; Egert, A.; Pieri, P.; Lecourieux, F.; Kappel, C.; Charon, J.; Gomes, E.; Keller, F.; Delrot, S.; Lecourieux, D. VvGOLS1 and VvHsfA2 are involved in the heat stress responses in grapevine berries. Plant Cell Physiol. 2012, 53, 1776–1792. [Google Scholar] [CrossRef] [PubMed]
- Rienth, M.; Torregrosa, L.; Luchaire, N.; Chatbanyong, R.; Lecourieux, D.; Kelly, M.T.; Romieu, C. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol. 2014, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Wang, J.F.; Cramer, G.; Dai, Z.W.; Duan, W.; Xu, H.G.; Wu, B.H.; Fan, P.G.; Wang, L.J.; Li, S.H. Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol. 2012, 12, 174. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-J.J.; Krenz, D.C.; Galvez, A.F.; de Lumen, B.O. Galactinol synthase (GS): Increased enzyme activity and levels of mRNA due to cold and desiccation. Plant Sci. 1998, 134, 11–20. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef]
- Orthen, B.; Popp, M.; Smirnoff, N. Hydroxyl radical scavenging properties of cyclitols. Proc. R. Soc. Edinb. Sect. B. Biol. Sci. 1994, 102, 269–272. [Google Scholar] [CrossRef]
- Saravitz, D.M.; Pharr, D.M.; Carter, T.E. Galactinol synthase activity and soluble sugars in developing seeds of four soybean genotypes. Plant Physiol. 1987, 83, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Brenac, P.; Horbowicz, M.; Downer, S.M.; Dickerman, A.M.; Smith, M.E.; Obendorf, R.L. Raffinose accumulation related to desiccation tolerance during maize (Zea mays L.) seed development and maturation. J. Plant Physiol. 1997, 150, 481–488. [Google Scholar] [CrossRef]
- Liu, H.L.; Dai, X.Y.; Xu, Y.Y.; Chong, K. Over-expression of OsUGE-1 altered raffinose level and tolerance to abiotic stress but not morphology in Arabidopsis. J. Plant Physiol. 2007, 164, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Janská, A.; Aprile, A.; Zamecnik, J.; Cattivelli, L.; Ovesna, J. Transcriptional responses of winter barley to cold indicate nucleosome remodelling as a specific feature of crown tissues. Funct. Integr. Genom. 2011, 11, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Sung, D.Y.; Guy, C.L. Roles of β-amylase and starch breakdown during temperatures stress. Physiol. Plant. 2006, 126, 120–128. [Google Scholar] [CrossRef]
- Kaplan, F.; Guy, C.L. β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004, 135, 1674–1684. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef]
- Liu, H.; Ma, Y.; Chen, N.; Guo, S.; Liu, H.; Guo, X.; Chong, K.; Xu, Y. Overexpression of stress-inducible OsBURP16, the beta subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 2014, 37, 1144–1158. [Google Scholar] [CrossRef]
- Sawicki, M.; Jacquens, L.; Baillieul, F.; Clément, C.; Vaillant-Gaveau, N.; Jacquard, C. Distinct regulation in inflorescence carbohydrate metabolism according to grapevine cultivars during floral development. Physiol. Plant. 2015, 154, 447–467. [Google Scholar] [CrossRef] [PubMed]
- Estornell, L.H.; Agusti, J.; Merelo, P.; Talón, M.; Tadeo, F.R. Elucidating mechanisms underlying organ abscission. Plant Sci. 2013, 199–200, 48–60. [Google Scholar] [CrossRef] [PubMed]
- González-Carranza, Z.H.; Lozoya-Gloria, E.; Roberts, J.A. Recent developments in abscission: Shedding light on the shedding process. Trends Plant Sci. 1998, 3, 10–14. [Google Scholar] [CrossRef]
- Taylor, J.E.; Whitelaw, C.A. Signals in abscission. New Phytol. 2001, 151, 323–340. [Google Scholar] [CrossRef]
- Orozco-Cárdenas, M.L.; Ryan, C.A. Polygalacturonase beta-subunit antisense gene expression in tomato plants leads to a progressive enhanced wound response and necrosis in leaves and abscission of developing flowers. Plant Physiol. 2003, 133, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G.; Stead, A.D. Abscission of flowers and floral parts. J. Exp. Bot. 1997, 48, 821–837. [Google Scholar] [CrossRef]
- Del Campillo, E.; Bennett, A.B. Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol. 1996, 111, 813–820. [Google Scholar] [CrossRef]
- Lashbrook, C.C.; Cai, S. Cell wall remodeling in Arabidopsis stamen abscission zones: Temporal aspects of control inferred from transcriptional profiling. Plant Signal. Behav. 2008, 3, 733–736. [Google Scholar] [CrossRef]
- Huglin, P.; Schneider, C. Biologie et Écologie de la Vigne, 2nd ed.; Lavoisier: Paris, France, 1998; p. 372. [Google Scholar]
- Zhu, Y.N.; Shi, D.Q.; Ruan, M.B.; Zhang, L.L.; Meng, Z.H.; Liu, J.; Yang, W.C. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e80218. [Google Scholar] [CrossRef]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Walter, M.H.; Grima-Pettenati, J.; Grand, C.; Boudet, A.M.; Lamb, C.J. Cinnamyl-alcohol dehydrogenase, a molecular marker specific for lignin synthesis: cDNA cloning and mRNA induction by fungal elicitor. Proc. Natl. Acad. Sci. USA 1988, 85, 5546–5550. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Jeong, J.C.; Lee, H.S.; Kwak, S.S. Comparative characterization of sweetpotato antioxidant genes from expressed sequence tags of dehydration-treated fibrous roots under different abiotic stress conditions. Mol. Biol. Rep. 2013, 40, 2887–2896. [Google Scholar] [CrossRef] [PubMed]
- Rajashekar, C.B.; Lafta, A. Cell-Wall Changes and Cell Tension in Response to Cold Acclimation and Exogenous Abscisic Acid in Leaves and Cell Cultures. Plant Physiol. 1996, 111, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef] [PubMed]
- Badowiec, A.; Swigonska, S.; Weidner, S. Changes in the protein patterns in pea (Pisum sativum L.) roots under the influence of long- and short-term chilling stress and post-stress recovery. Plant Physiol. Biochem. 2013, 71, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Badowiec, A.; Weidner, S. Proteomic changes in the roots of germinating Phaseolus vulgaris seeds in response to chilling stress and post-stress recovery. J. Plant Physiol. 2014, 171, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cezard, L.; Le Bris, P.; Borrega, N.; Herve, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef]
- Olenichenko, N.A.; Zagoskina, N.V. Response of winter wheat to cold: Production of phenolic compounds and L-phenylalanine ammonia-lyase activity. Appl. Biochem. Microbiol. 2005, 41, 600–603. [Google Scholar] [CrossRef]
- Lallemand, B.; Erhardt, M.; Heitz, T.; Legrand, M. Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol. 2013, 162, 616–625. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- De Azevedo Souza, C.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, S.M.; Haughn, G.W.; Kombrink, E.; Douglas, C.J. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids--biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yu, Y.; Ding, J.; Hua, Z.; Wang, Y. Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 2010, 231, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Kodan, A.; Kuroda, H.; Sakai, F. A stilbene synthase from Japanese red pine (Pinus densiflora): Implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 3335–3339. [Google Scholar] [CrossRef] [PubMed]
- Lebon, G.; Duchene, E.; Brun, O.; Clément, C. Phenology of flowering and starch accumulation in grape (Vitis vinifera L.) cuttings and vines. Ann. Bot. 2005, 95, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.H.; Dudoit, S.; Luu, P.; Lin, D.M.; Peng, V.; Ngai, J.; Speed, T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002, 30, e15. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Muller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
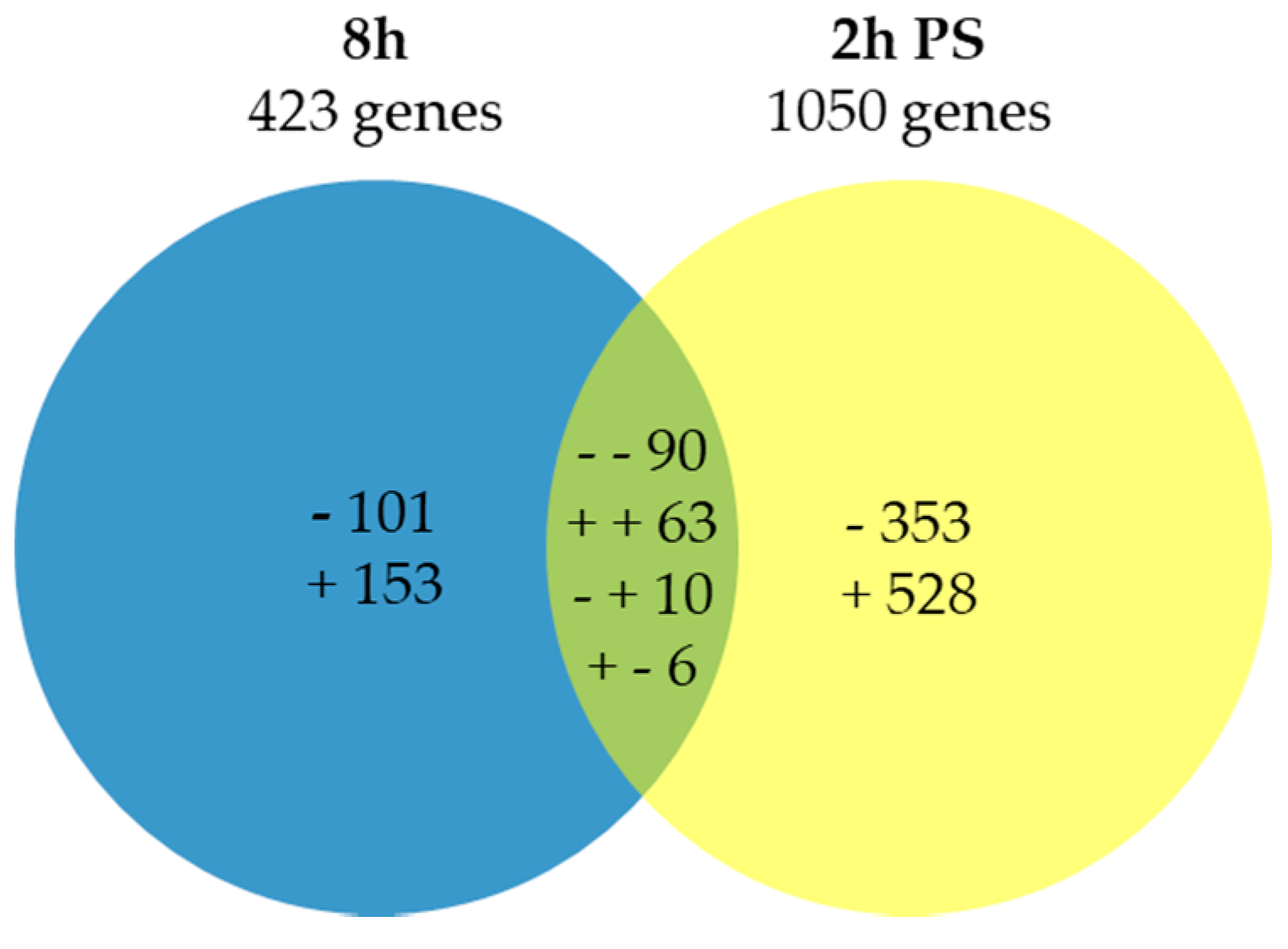






| 8 h | ||
|---|---|---|
| BIN | Up-Regulated Process | Adjusted p-Value |
| 10 | cell wall | 1.7110 × 10−4 |
| 10.7 | cell wall.modification | 5.3204 × 10−4 |
| 16.8 | secondary metabolism.flavonoids | 6.3979 × 10−3 |
| 16.8.3 | secondary metabolism.flavonoids.dihydroflavonols | 7.0367 × 10−3 |
| 16.8.3.1 | secondary metabolism.flavonoids.dihydroflavonols, .dihydrokaempferol 4-reductase | 5.3735 × 10−3 |
| 20 | stress | 2.2949 × 10−8 |
| 20.1 | stress.biotic | 3.8506 × 10−3 |
| 20.1.7 | stress.biotic.PR-proteins | 6.15 × 10−3 |
| 20.2 | stress.abiotic | 8.0636 × 10−6 |
| 20.2.1 | stress.abiotic.heat | 3.0648 × 10−7 |
| 24 | Biodegradation of Xenobiotics | 2.1821 × 10−3 |
| 27 | RNA | 4.0490 × 10−4 |
| 27.1.1 | RNA.processing.splicing | 8.2208 × 10−3 |
| 27.3 | RNA regulation of transcription | 1.5298 × 10−4 |
| 27.3.3 | RNA regulation of transcription.AP2_EREBP, APETALA2_Ethylene-responsive element binding protein family | 9.5059 × 10−13 |
| 27.3.66 | RNA regulation of transcription.Psudo ARR transcription factor family | 4.7432 × 10−7 |
| 29 | protein | 3.4384 × 10−3 |
| 29.5.1 | protein degradation.subtilases | 9.6717 × 10−6 |
| 30.1 | signaling in sugar and nutrient physiology | 5.8130 × 10−5 |
| 30.3 | signaling.calcium | 1.3157 × 10−3 |
| 34.3 | transport.amino acids | 5.32 × 10−4 |
| BIN | Down-Regulated Process | Adjusted p-Value |
| 3 | minor CHO metabolism | 2.8967 × 10−3 |
| 3.1 | minor CHO metabolism.raffinose family | 2.9243 × 10−5 |
| 3.1.1 | minor CHO metabolism.raffinose family.galactinol synthases | 2.4621 × 10−6 |
| 3.1.1.2 | minor CHO metabolism.raffinose family.galactinol synthases.putative | 2.4621 × 10−6 |
| 5.3 | Fermentation.ADH | 5.7860 × 10−3 |
| 8.3 | TCA_org transformation.carbonic anhydrases | 1.2604 × 10−4 |
| 10 | cell wall | 2.2613 × 10−4 |
| 10.6 | cell walldegradation | 5.4979 × 10−4 |
| 10.6.3 | cell wall.degradation.pectate lyases and polygalacturonases | 1.3581 × 10−5 |
| 10.7 | cell wall.modification | 3.0702× 10−3 |
| 16 | secondary metabolism | 3.2141 × 10−12 |
| 16.1 | secondary metabolism.isoprenoids | 4.1006 × 10−13 |
| 16.1.4 | secondary metabolism.isoprenoids.carotenoids | 4.4294 × 10−3 |
| 16.1.5 | secondary metabolism.isoprenoids.terpenoids | 3.7027 × 10−16 |
| 16.8 | secondary metabolism.flavonoids | 1.7181 × 10−4 |
| 16.8.1 | secondary metabolism.flavonoids.anthocyanins | 2.9899 × 10−5 |
| 16.8.1.12 | secondary metabolism.flavonoids, anthocyanins.anthocyanidin 3-O-glucosyltransferase | 2.7661 × 10−4 |
| 16.8.1.21 | secondary metabolism.flavonoids, anthocyanins.anthocyanin 5-aromatic acyltransferase | 1.4022 × 10−3 |
| 17.8 | hormone metabolism.salicylic acid | 2.0278 × 10−3 |
| 17.8.1 | hormone metabolism.salicylic acid.synthesis-degradation | 1.8638 × 10−3 |
| 19 | tetrapyrrole synthesis | 2.5726 × 10−3 |
| 27.3.26 | RNA regulation of transcription.MYB-related transcription factor family | 6.6359 × 10−6 |
| 27.3.7 | RNA regulation of transcription.C2C2(Zn) CO-like, Constans-like zinc finger family | 2.0362 × 10−8 |
| 30.11 | signaling light | 3.7892 × 10−3 |
| 34 | transport | 6.5478 × 10−5 |
| 34.19 | transport.major intrinsic proteins | 2.0278 × 10−3 |
| 34.4 | transport.nitrate | 5.7860 × 10−3 |
| 34.6 | transport.sulphate | 1.9185 × 10−4 |
| 2 h PS | ||
|---|---|---|
| BIN | Up-Regulated Process | Adjusted p-Value |
| 1.3.1 | PS.calvin cycle.rubisco large subunit | 2.4217 × 10−5 |
| 3 | minor CHO metabolism | 2.4623 × 10−3 |
| 3.1 | minor CHO metabolism.raffinose family | 6.9210 × 10−7 |
| 3.1.1 | minor CHO metabolism.raffinose family.galactinol synthases | 7.6070 × 10−6 |
| 3.1.1.2 | minor CHO metabolism.raffinose family.galactinol synthases.putative | 7.6070 × 10−6 |
| 11.9.3.5 | lipid metabolism.lipid degradation.lysophospholipases.phosphoinositide phospholipase C | 4.3982 × 10−3 |
| 16.2 | secondary metabolism.phenylpropanoids | 1.3950 × 10−3 |
| 16.2.1 | secondary metabolism.phenylpropanoids.lignin biosynthesis | 1.4851 × 10−4 |
| 16.2.1.10 | secondary metabolism.phenylpropanoids.lignin biosynthesis.CAD | 5.2999 × 10−6 |
| 16.8.3 | secondary metabolism.flavonoids.dihydroflavonols | 4.3804 × 10−3 |
| 17 | hormone metabolism | 1.2461 × 10−3 |
| 17.1.3 | hormone metabolism.abscisic acid.induced-regulated-responsive-activated | 1.5786 × 10−3 |
| 17.8 | hormone metabolism.salicylic acid | 8.5038 × 10−4 |
| 17.8.1 | hormone metabolism.salicylic acid.synthesis-degradation | 7.4185 × 10−4 |
| 20 | stress | 3.7621 × 10−7 |
| 20.1 | stress.biotic | 1.2488 × 10−8 |
| 20.1.2 | stress.biotic.receptors | 1.8208 × 10−3 |
| 20.1.3 | stress.biotic.signaling | 2.9375 × 10−8 |
| 20.1.7 | stress.biotic.PR-proteins | 4.1661 × 10−3 |
| 20.2.3 | stress.abiotic.drought_salt | 4.8258 × 10−4 |
| 26.12 | misc.peroxidases | 2.0486 × 10−3 |
| 26.9 | misc.glutathione S transferases | 1.5046 × 10−3 |
| 27.3 | RNA.regulation of transcription | 7.5119 × 10−4 |
| 27.3.22 | RNA.regulation of transcription.HB,Homeobox transcription factor family | 5.9729 × 10−5 |
| 27.3.27 | RNA.regulation of transcription.NAC domain transcription factor family | 1.6861 × 10−4 |
| 27.3.6 | RNA.regulation of transcription.bHLH,Basic Helix-Loop-Helix family | 7.6192 × 10−4 |
| 28.1.3 | DNA.synthesis_chromatin structure.histone | 1.2067 × 10−5 |
| 29.2 | protein.synthesis | 2.6618 × 10−3 |
| 29.5.11 | protein.degradation.ubiquitin | 2.3331 × 10−12 |
| 29.5.9 | protein.degradation.AAA type | 1.3294 × 10−3 |
| 30 | signaling | 2.6317 × 10−4 |
| 30.2 | signaling.receptor kinases | 6.4535 × 10−4 |
| 30.2.20 | signaling.receptor kinases.wheat LRK10 like | 1.7662 × 10−4 |
| 30.2.99 | signaling.receptor kinases.misc | 9.1461 × 10−4 |
| 30.3 | signaling.calcium | 5.1743 × 10−4 |
| 34.13 | transport.peptides and oligopeptides | 1.2525 × 10−4 |
| 34.19 | transport.Major Intrinsic Proteins | 9.0675 × 10−5 |
| 34.19.1 | transport.Major Intrinsic Proteins.PIP | 5.5615 × 10−4 |
| BIN | Down-Regulated Process | Adjusted p-Value |
| 1 | PS | 4.5585 × 10−5 |
| 1.1 | PS.lightreaction | 2.8822 × 10−5 |
| 1.1.1 | PS.lightreaction.photosystem II | 3.0443 × 10−8 |
| 1.1.1.1 | PS.lightreaction.photosystem II.LHC-II | 4.7652 × 10−15 |
| 2.1.2 | major CHO metabolism.synthesis.starch | 4.1047 × 10−3 |
| 2.2.1.3.3 | major CHO metabolism.degradation.sucrose.invertases.vacuolar | 2.5554 × 10−4 |
| 10 | cell wall | 1.7409 × 10−20 |
| 10.2 | cell wall.cellulose synthesis | 8.9264 × 10−10 |
| 10.2.1 | cell wall.cellulose synthesis.cellulose synthase | 1.7420 × 10−6 |
| 10.6 | cell wall.degradation | 4.2186 × 10−10 |
| 10.6.3 | cell wall.degradation.pectate lyases and polygalacturonases | 1.6981 × 10−7 |
| 10.7 | cell wall.modification | 8.4556 × 10−9 |
| 11.1 | lipid metabolism.FA synthesis and FA elongation | 2.8132 × 10−3 |
| 13 | amino acid metabolism | 1.3271 × 10−5 |
| 13.1 | amino acid metabolism.synthesis | 1.1606 × 10−5 |
| 13.1.2 | amino acid metabolism.synthesis.glutamate family | 1.2995 × 10−3 |
| 13.1.5.1 | amino acid metabolism.synthesis.serine-glycine-cysteine group.serine | 3.6731 × 10−3 |
| 13.1.5.1.1 | amino acid metabolism.synthesis.serine-glycine-cysteine group.serine.phosphoglycerate dehydrogenase | 1.5008 × 10−3 |
| 16 | secondary metabolism | 2.0116 × 10−19 |
| 16.1 | secondary metabolism.isoprenoids | 1.0312 × 10−7 |
| 16.1.5 | secondary metabolism.isoprenoids.terpenoids | 3.5231 × 10−11 |
| 16.2 | secondary metabolism.phenylpropanoids | 7.0813 × 10−12 |
| 16.2.1 | secondary metabolism.phenylpropanoids.lignin biosynthesis | 1.2317 × 10−12 |
| 16.2.1.1 | secondary metabolism.phenylpropanoids.lignin biosynthesis.PAL | 3.0248 × 10−20 |
| 16.8 | secondary metabolism.flavonoids | 2.2763 × 10−6 |
| 16.8.2 | secondary metabolism.flavonoids.chalcones | 5.3110 × 10−5 |
| 16.8.2.1 | secondary metabolism.flavonoids.chalcones.naringenin-chalcone synthase | 1.3583 × 10−4 |
| 16.8.2.3.1 | secondary metabolism.flavonoids.chalcones.pterostilbene synthase | 4.0714 × 10−6 |
| 16.8.3 | secondary metabolism.flavonoids.dihydroflavonols | 4.2622 × 10−3 |
| 20.1 | stress.biotic | 4.9962 × 10−5 |
| 20.2 | stress.abiotic | 1.8297 × 10−5 |
| 20.2.1 | stress.abiotic.heat | 1.7390 × 10−7 |
| 29 | protein | 1.5050 × 10−3 |
| 29.5.1 | protein.degradation.subtilases | 3.8388 × 10−3 |
| 29.5.11 | protein.degradation.ubiquitin | 1.8703 × 10−3 |
| 31 | cell | 2.6211 × 10−3 |
| 33.99 | development.unspecified | 1.7360 × 10−3 |
| 34 | transport | 4.7864 × 10−12 |
| 34.10 | transport.nucleotides | 1.0332 × 10−3 |
| 34.4 | transport.nitrate | 1.9522 × 10−3 |
| 34.6 | transport.sulphate | 1.0111 × 10−4 |
| 34.99 | transport.misc | 3.4894 × 10−6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawicki, M.; Rondeau, M.; Courteaux, B.; Rabenoelina, F.; Guerriero, G.; Gomès, E.; Soubigou-Taconnat, L.; Balzergue, S.; Clément, C.; Ait Barka, E.; et al. On a Cold Night: Transcriptomics of Grapevine Flower Unveils Signal Transduction and Impacted Metabolism. Int. J. Mol. Sci. 2019, 20, 1130. https://doi.org/10.3390/ijms20051130
Sawicki M, Rondeau M, Courteaux B, Rabenoelina F, Guerriero G, Gomès E, Soubigou-Taconnat L, Balzergue S, Clément C, Ait Barka E, et al. On a Cold Night: Transcriptomics of Grapevine Flower Unveils Signal Transduction and Impacted Metabolism. International Journal of Molecular Sciences. 2019; 20(5):1130. https://doi.org/10.3390/ijms20051130
Chicago/Turabian StyleSawicki, Mélodie, Marine Rondeau, Barbara Courteaux, Fanja Rabenoelina, Gea Guerriero, Eric Gomès, Ludivine Soubigou-Taconnat, Sandrine Balzergue, Christophe Clément, Essaïd Ait Barka, and et al. 2019. "On a Cold Night: Transcriptomics of Grapevine Flower Unveils Signal Transduction and Impacted Metabolism" International Journal of Molecular Sciences 20, no. 5: 1130. https://doi.org/10.3390/ijms20051130