Comparative Transcriptome Combined with Proteome Analyses Revealed Key Factors Involved in Alfalfa (Medicago sativa) Response to Waterlogging Stress
Abstract
1. Introduction
2. Results
2.1. Physiological Response to Waterlogging
2.2. Transcriptome Sequencing and Assembly
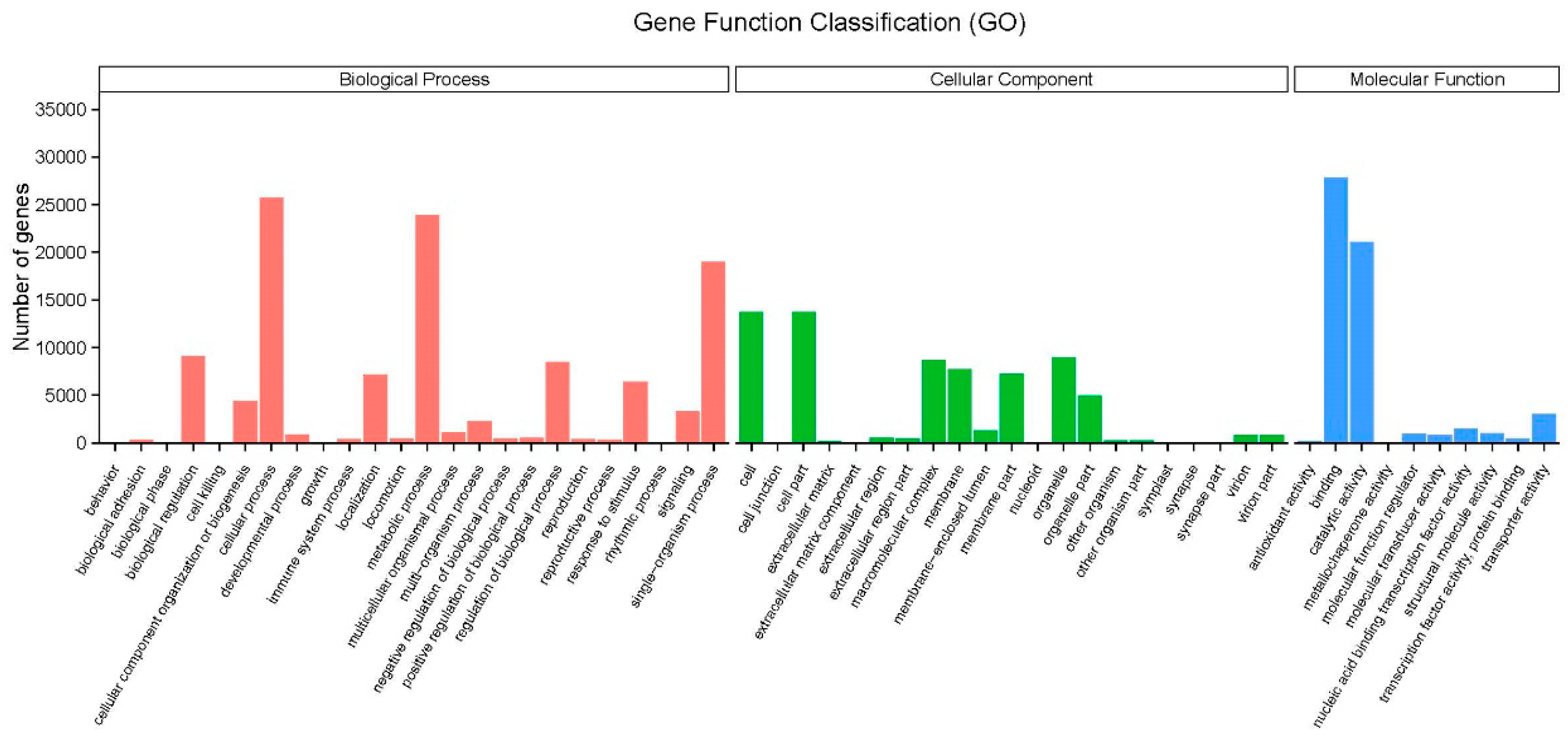
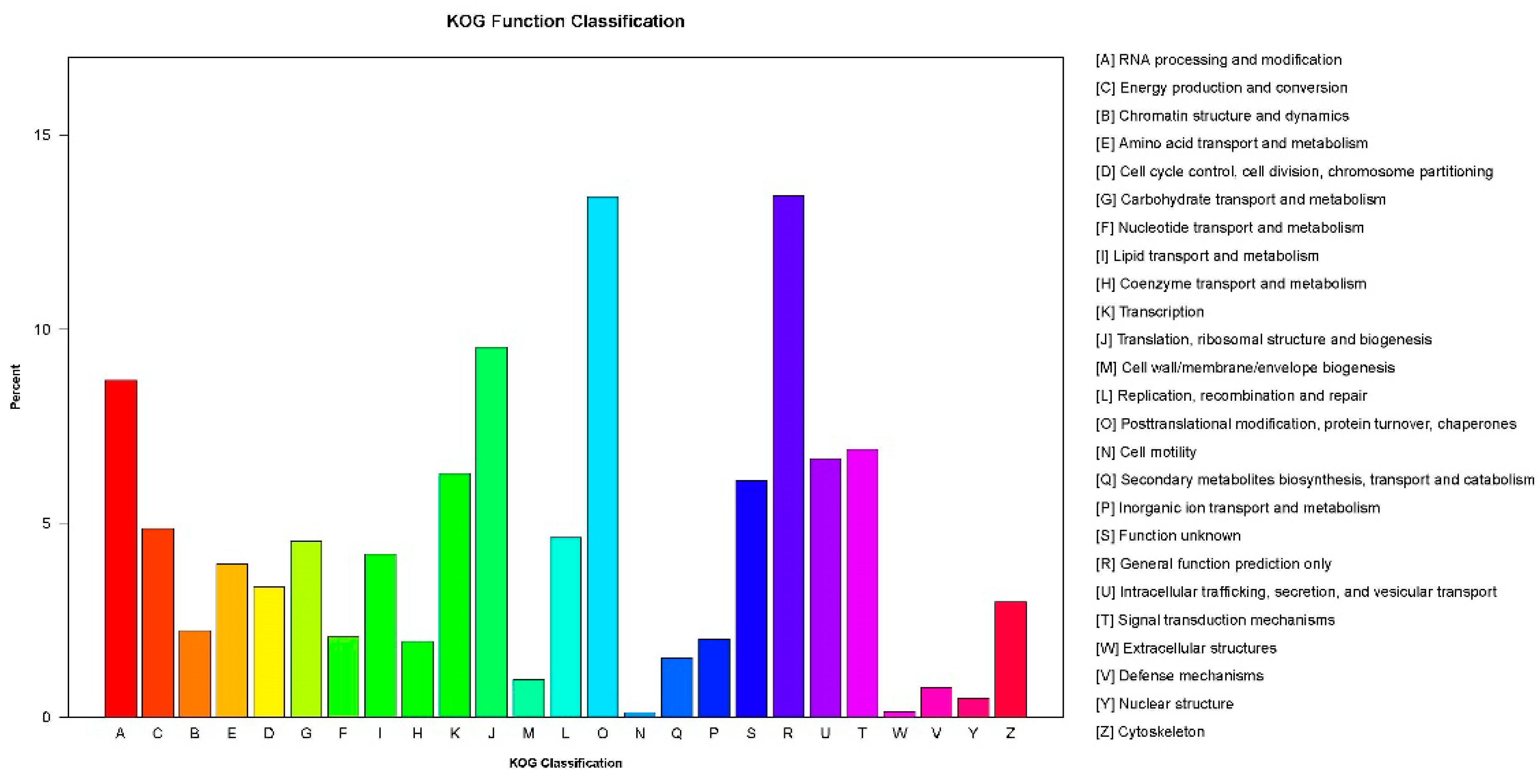
2.3. Gene ontology (GO) and KOG Classification
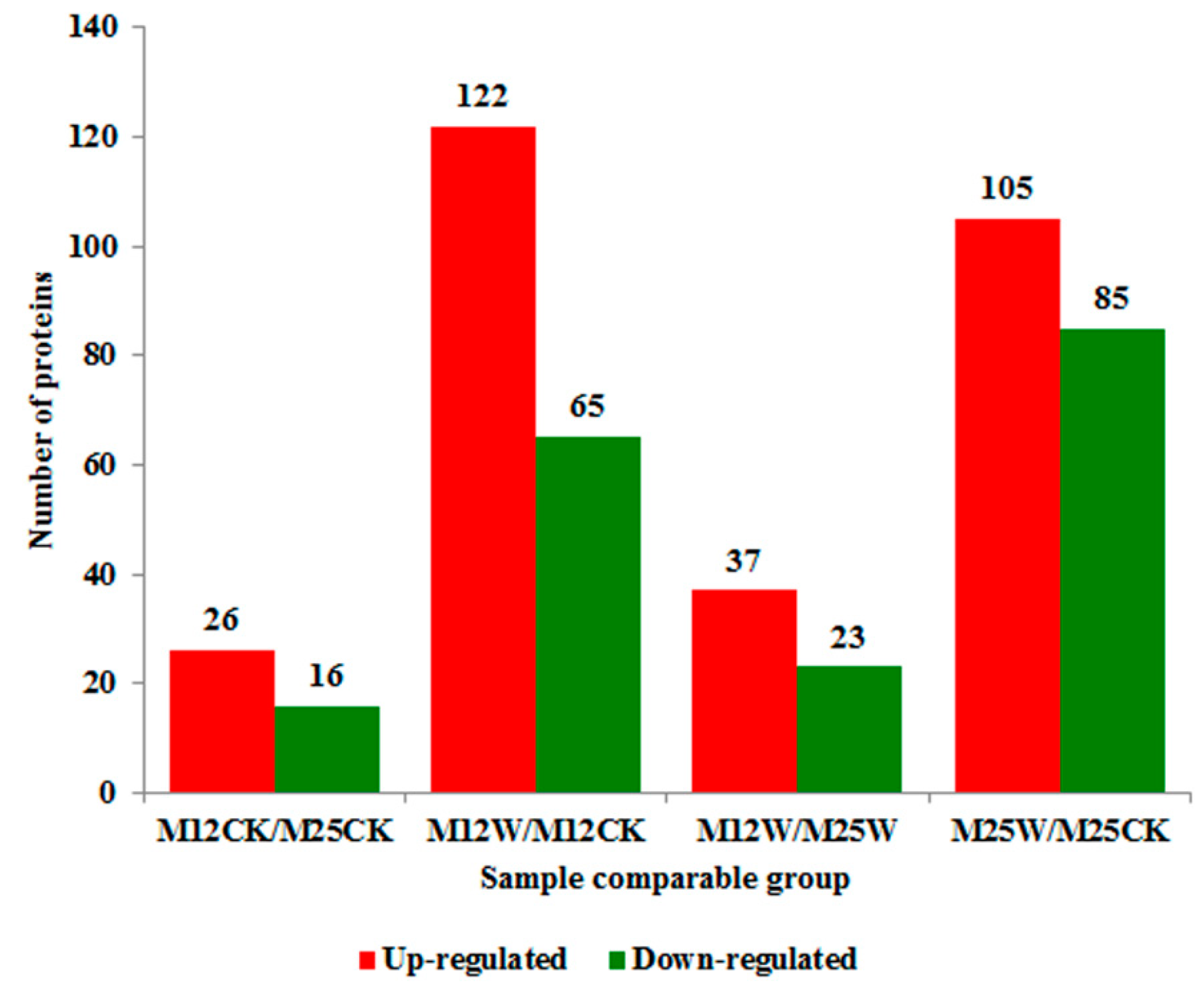
2.4. Differentially Abundant Transcripts (DATs) Analysis
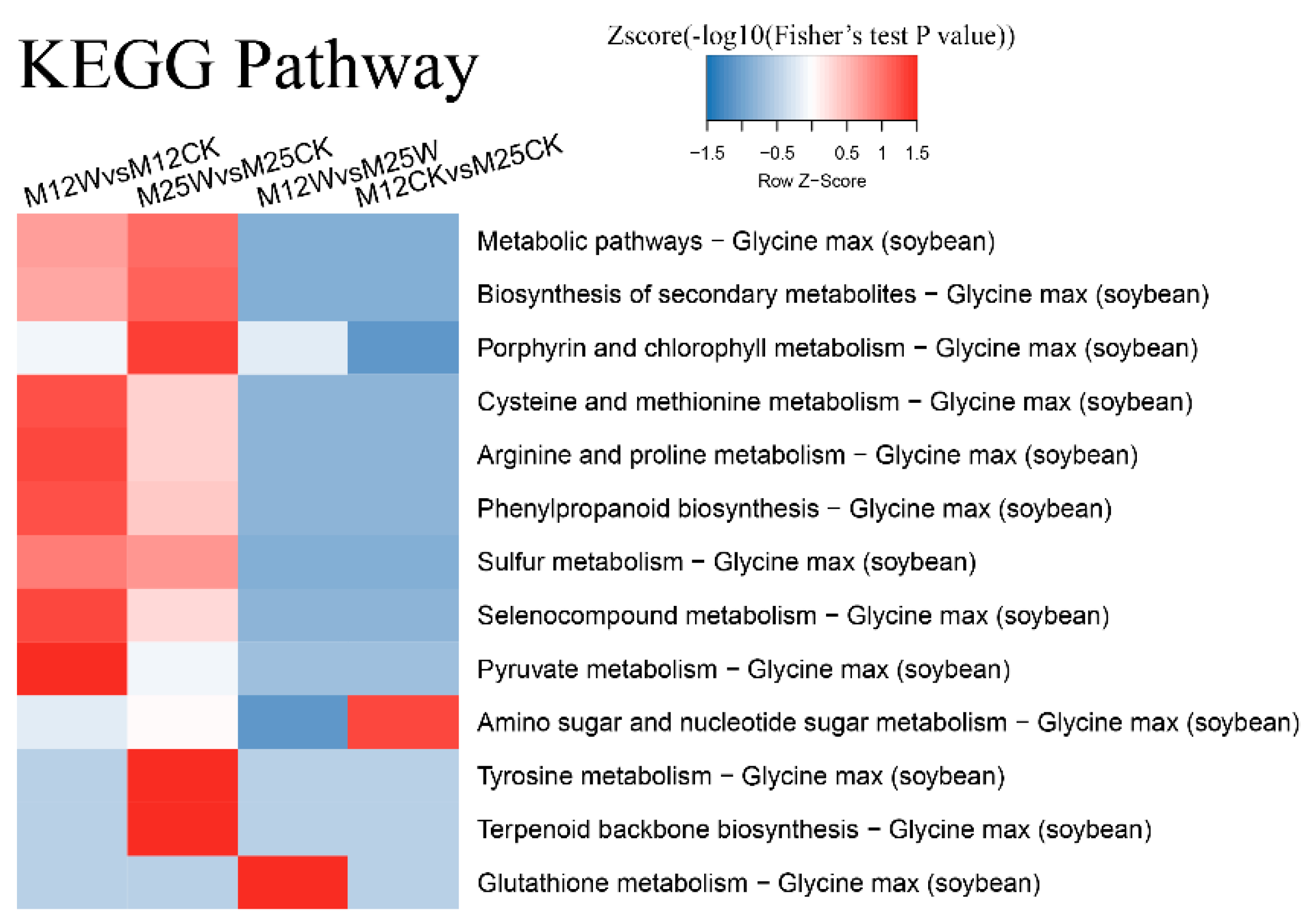
2.5. Function Annotation of DATs Using the KEGG Database
2.6. Proteome-Wide Analysis of Differentially Abundant Proteins by Waterlogging Treatment
2.7. Gene Ontology (GO) Analysis of DAPs
2.8. KEGG Analysis of DAPs
2.9. Transcriptomes and Proteomics Crosstalk Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Waterlogging Treatment
4.2. Leaf Chlorophyll Content, Maximum Quantum Yield of Photosystem II Efficiency (Fv/Fm), and Net Photosynthetic Rate (Pn) Determination
4.3. RNA Preparation, Sequencing, and Data Analysis
4.4. Protein Extraction and Trypsin Digestion
4.5. iTRAQ Labeling
4.6. HPLC Fractionation and LC-MS/MS Analysis
4.7. Database Search
4.8. Validation of Differentially Expressed Unigenes and Proteins by Quantitative Real-Time PCR (qRT-PCR) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bagavathiannan, M.V.; Van Acker, R.C. The biology and ecology of feral alfalfa (Medicago sativa L.) and its implications for novel trait confinement in North America. Crit. Rev. Plant Sci. 2009, 28, 69–87. [Google Scholar] [CrossRef]
- Barta, A.; Sulc, R. Interaction between waterlogging injury and irradiance level in alfalfa. Crop Sci. 2002, 42, 1529–1534. [Google Scholar] [CrossRef]
- Breazeale, D.; Neufeld, J.; Myer, G.; Davison, J. Feasibility of subsurface drip irrigation for alfalfa. J. ASFMRA 2000, 1, 58–63. [Google Scholar]
- Samac, D.A.; Jung, H.; Lamb, J. Development of alfalfa (Medicago sativa L.) as a feedstock for production of ethanol and other bioproducts. Chem. Ind.-N. Y.-Marcel Dekk. 2006, 112, 79. [Google Scholar]
- Humphries, A.W.; Auricht, G. Breeding lucerne for Australias southern dryland cropping environments. Aust. J. Agric. Res. 2001, 52, 153–169. [Google Scholar] [CrossRef]
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef]
- Dat, J.F.; Capelli, N.; Folzer, H.; Bourgeade, P.; Badot, P.M. Sensing and signalling during plant flooding. Plant Physiol. Biochem. 2004, 42, 273–282. [Google Scholar] [CrossRef]
- Drew, M.C. Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Ann. Rev. Plant Biol. 1997, 48, 223–250. [Google Scholar] [CrossRef]
- Christianson, J.A.; Llewellyn, D.J.; Dennis, E.S.; Wilson, I.W. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2009, 51, 21–37. [Google Scholar] [CrossRef]
- Christianson, J.A.; Llewellyn, D.J.; Dennis, E.S.; Wilson, I.W. Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species. Plant Signal. Behav. 2010, 5, 1006–1009. [Google Scholar] [CrossRef]
- Nanjo, Y.; Maruyama, K.; Yasue, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Komatsu, S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol. Biol. 2011, 77, 129–144. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Huang, S.N.; Mo, Z.H.; Xuan, J.P.; Jia, X.D.; Wang, G.; Guo, Z.R. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol. Breed. 2015, 35, 208. [Google Scholar] [CrossRef]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef]
- Klok, E.J.; Wilson, I.W.; Wilson, D.; Chapman, S.C.; Ewing, R.M.; Somerville, S.C.; Peacock, W.J.; Dolferus, R.; Dennis, E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 2002, 14, 2481–2494. [Google Scholar] [CrossRef]
- Liu, F.; VanToai, T.; Moy, L.P.; Bock, G.; Linford, L.D.; Quackenbush, J. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 2005, 137, 1115–1129. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, D.G.; Lee, S.H.; Kang, K.Y.; Bahk, J.D.; Choi, M.S.; Lee, I.J.; Renaut, J.; Lee, B.H. A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiol. Plant. 2007, 131, 555–570. [Google Scholar] [CrossRef]
- Komatsu, S.; Kamal, A.H.; Hossain, Z. Wheat proteomics: Proteome modulation and abiotic stress acclimation. Front. Plant Sci. 2014, 5, 684. [Google Scholar] [CrossRef]
- Komatsu, S.; Shirasaka, N.; Sakata, K. ‘Omics’ techniques for identifying flooding–response mechanisms in soybean. J. Proteom. 2013, 93, 169–178. [Google Scholar] [CrossRef]
- Wang, X.; Huang, M.; Zhou, Q.; Cai, J.; Dai, T.; Cao, W.; Jiang, D. Physiological and proteomic mechanisms of waterlogging priming improves tolerance to waterlogging stress in wheat (Triticum aestivum L.). Environ. Exp. Bot. 2016, 132, 175–182. [Google Scholar] [CrossRef]
- Yu, F.; Han, X.; Geng, C.; Zhao, Y.; Zhang, Z.; Qiu, F. Comparative proteomic analysis revealing the complex network associated with waterlogging stress in maize (Zea mays L.) seedling root cells. Proteomics 2015, 15, 135–147. [Google Scholar] [CrossRef]
- Mujer, C.V.; Rumpho, M.E.; Lin, J.J.; Kennedy, R.A. Constitutive and inducible aerobic and anaerobic stress proteins in the Echinochloa complex and rice. Plant Physiol. 1993, 101, 217–226. [Google Scholar] [CrossRef]
- Sachs, M.M.; Freeling, M.; Okimoto, R. The anaerobic proteins of maize. Cell 1980, 20, 761–767. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, D.; Zheng, Y. Transcriptional and post-transcriptional regulation of gene expression in submerged root cells of maize. Plant Signal. Behav. 2009, 4, 132–135. [Google Scholar] [CrossRef][Green Version]
- Alam, I.; Lee, D.G.; Kim, K.H.; Park, C.H.; Sharmin, S.A.; Lee, H.; Oh, K.W.; Yun, B.W.; Lee, B.H. Proteome analysis of soybean roots under waterlogging stress at an early vegetative stage. J. Biosci. 2010, 35, 49–62. [Google Scholar] [CrossRef]
- Narsai, R.; Rocha, M.; Geigenberger, P.; Whelan, J.; van Dongen, J.T. Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol. 2011, 190, 472–487. [Google Scholar] [CrossRef]
- Das, A.; Uchimiya, H. Oxygen stress and adaptation of a semi-aquatic plant: Rice (Oryza sativa). J. Plant Res. 2002, 115, 315–320. [Google Scholar] [CrossRef]
- Guglielminetti, L.; Perata, P.; Alpi, A. Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol. 1995, 108, 735–741. [Google Scholar] [CrossRef]
- Harper, J.F. Dissecting calcium oscillators in plant cells. Trends Plant Sci. 2001, 6, 395–397. [Google Scholar] [CrossRef]
- Sedbrook, J.C.; Kronebusch, P.J.; Borisy, G.G.; Trewavas, A.J.; Masson, P.H. Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiol. 1996, 111, 243–257. [Google Scholar] [CrossRef]
- Subbaiah, C.C.; Zhang, J.; Sachs, M.M. Involvement of intracellular calcium in Anaerobic gene expression and survival of maize seedlings. Plant Physiol. 1994, 105, 369–376. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef]
- Lee, K.W.; Chen, P.W.; Lu, C.A.; Chen, S.; Ho, T.H.D.; Yu, S.M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009, 2, ra61. [Google Scholar] [CrossRef]
- Ye, N.H.; Wang, F.Z.; Shi, L.; Chen, M.X.; Cao, Y.Y.; Zhu, F.Y.; Wu, Y.Z.; Xie, L.J.; Liu, T.Y.; Su, Z.Z.; et al. Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J. 2018, 94, 612–625. [Google Scholar] [CrossRef]
- Zhao, N.; Li, C.; Yan, Y.; Cao, W.; Song, A.; Wang, H.; Chen, S.; Jiang, J.; Chen, F. Comparative transcriptome analysis of waterlogging-sensitive and waterlogging-tolerant Chrysanthemum morifolium cultivars under waterlogging stress and reoxygenation conditions. Int. J. Mol. Sci. 2018, 19, E1455. [Google Scholar] [CrossRef]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy metal tolerance in plants: Role of transcriptomics, proteomics, metabolomics, and Ionomics. Front. Plant Sci. 2016, 6, 1143. [Google Scholar] [CrossRef]
- Zhu, H.; Ai, H.; Cao, L.; Sui, R.; Ye, H.; Du, D.; Sun, J.; Yao, J.; Chen, K.; Chen, L. Transcriptome analysis providing novel insights for Cd-resistant tall fescue responses to Cd stress. Ecotoxicol. Environ. Saf. 2018, 160, 349–356. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, sR106. [Google Scholar] [CrossRef]
- Storey, J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Cui, W.; Pan, J.; Xie, Y.; Wang, J.; Shen, W. Proteomic analysis provides insights into the molecular bases of hydrogen gas-induced cadmium resistance in Medicago sativa. J. Proteom. 2017, 152, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, X.; Zhang, Z.; Liu, N.; Li, D.; Hu, L. Melatonin improved waterlogging tolerance in alfalfa (Medicago sativa) by reprogramming polyamine and ethylene metabolism. Front. Plant Sci. 2019, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Sample | Raw Reads | Clean Reads | Clean Bases | Error (%) | Q 20 (%) | Q 30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| M12_W | 46788858 | 44893272 | 6.73G | 0.02 | 96.00 | 90.30 | 41.61 |
| M12_CK | 53839716 | 51414672 | 7.71G | 0.02 | 95.89 | 90.07 | 42.21 |
| M25_W | 50605680 | 48385890 | 7.26G | 0.02 | 95.84 | 90.01 | 41.42 |
| M25_CK | 53716242 | 51239418 | 7.69G | 0.02 | 95.70 | 89.65 | 42.09 |
| Category | Transcripts | Genes |
|---|---|---|
| 200–500 bp | 105,740 | 39,418 |
| 500–1000 bp | 36,445 | 34,759 |
| 1000–2000 bp | 25,605 | 25,570 |
| >2000 bp | 12,717 | 12,717 |
| Total | 180,507 | 112,464 |
| Min Length | 201 | 201 |
| Mean Length | 726 | 995 |
| Median Length | 405 | 681 |
| Max Length | 15,720 | 15,720 |
| N50 | 1196 | 1448 |
| N90 | 283 | 456 |
| Total Nucleotides | 131,136,850 | 111,915,817 |
| Pathway Term | Rich Factor | q Value | Gene Number |
|---|---|---|---|
| M12_W vs. M12_CK | |||
| Photosynthesis-antenna proteins | 0.4 | 3.94 × 10−8 | 10 |
| Arginine and proline metabolism | 0.080882353 | 3.52 × 10−3 | 11 |
| alpha-Linolenic acid metabolism | 0.084745763 | 3.52 × 10−3 | 10 |
| Nitrogen metabolism | 0.116666667 | 4.51 × 10−3 | 7 |
| Photosynthesis | 0.067961165 | 6.70 × 10−2 | 7 |
| Valine, leucine and isoleucine degradation | 0.058333333 | 1.20 × 10−1 | 7 |
| Carotenoid biosynthesis | 0.065789474 | 1.67 × 10−1 | 5 |
| Cysteine and methionine metabolism | 0.044117647 | 1.67 × 10−1 | 9 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 0.076923077 | 1.67 × 10−1 | 4 |
| Porphyrin and chlorophyll metabolism | 0.057471264 | 1.77 × 10−1 | 5 |
| Lysine degradation | 0.056818182 | 1.77 × 10−1 | 5 |
| Tryptophan metabolism | 0.055555556 | 1.77 × 10−1 | 5 |
| Isoquinoline alkaloid biosynthesis | 0.085714286 | 1.77 × 10−1 | 3 |
| Plant-pathogen interaction | 0.031078611 | 1.77 × 10−1 | 17 |
| Glutathione metabolism | 0.040816327 | 1.77 × 10−1 | 8 |
| Carbon fixation in photosynthetic organisms | 0.047244094 | 1.77 × 10−1 | 6 |
| Ascorbate and aldarate metabolism | 0.052083333 | 1.77 × 10−1 | 5 |
| Limonene and pinene degradation | 0.069767442 | 2.22 × 10−1 | 3 |
| Galactose metabolism | 0.040540541 | 2.67 × 10−1 | 6 |
| Glycerolipid metabolism | 0.039735099 | 2.67 × 10−1 | 6 |
| M25_W vs. M25_CK | |||
| Photosynthesis | 0.368932039 | 4.79 × 10−20 | 38 |
| Photosynthesis—antenna proteins | 0.68 | 3.55 × 10−12 | 17 |
| Carbon fixation in photosynthetic organisms | 0.204724409 | 5.17 × 10−9 | 26 |
| Nitrogen metabolism | 0.233333333 | 1.75 × 10−5 | 14 |
| Glyoxylate and dicarboxylate metabolism | 0.141732283 | 2.25 × 10−4 | 18 |
| Valine, leucine and isoleucine degradation | 0.141666667 | 3.28 × 10−4 | 17 |
| Arginine and proline metabolism | 0.132352941 | 3.68 × 10− | 18 |
| Porphyrin and chlorophyll metabolism | 0.16091954 | 3.79 × 10−4 | 14 |
| Fatty acid degradation | 0.094339623 | 3.23 × 10−2 | 15 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 0.11 | 3.54 × 10−2 | 11 |
| Cysteine and methionine metabolism | 0.083333333 | 4.67 × 10−2 | 17 |
| Ascorbate and aldarate metabolism | 0.104166667 | 6.19 × 10−2 | 10 |
| Pentose phosphate pathway | 0.093023256 | 6.19 × 10−2 | 12 |
| Tryptophan metabolism | 0.1 | 9.74 × 10−2 | 9 |
| Carotenoid biosynthesis | 0.105263158 | 1.0 × 10−1 | 8 |
| Glycine, serine and threonine metabolism | 0.077419355 | 1.63 × 10−1 | 12 |
| Tyrosine metabolism | 0.093023256 | 1.63 × 10−1 | 8 |
| Alanine, aspartate and glutamate metabolism | 0.085714286 | 1.73 × 10−1 | 9 |
| Peroxisome | 0.069444444 | 1.73 × 10−1 | 15 |
| beta-Alanine metabolisma | 0.075757576 | 2.33 × 10−1 | 10 |
| Protein Accession | Protein Description | MW (kDa) | M12W/M12CK Ratio | M12W/M12CK p-Value | Protein LOG2 M12W/M12CK | Regulation | Transcription Log2 M12_W/M12_CK | p adj | Regulation | Type |
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster-1252.18690_orf1 | Unknown | 42.266 | 1.816 | 5.1953 × 10−3 | 0.860764203 | Up | 1.7749 | 9.0263 × 10−7 | Up | Up-Up |
| Cluster-1252.20320_orf1 | Probable mannitol dehydrogenase OS = Medicago sativa GN=CAD1 PE=1 SV=1 | 39.526 | 1.414 | 2.8745 × 10−2 | 0.49978212 | Up | 1.664 | 2.132 × 10−4 | Up | Up-Up |
| Cluster-1252.24948_orf1 | Acidic endochitinase OS=Cicer arietinum PE=2 SV=1 | 33.034 | 2.91 | 5.1816 × 10−3 | 1.541019153 | Up | 3.0386 | 4.4982 × 10−18 | Up | Up-Up |
| Cluster-1252.25469_orf1 | Probable glutathione S-transferase OS=Glycine max GN=HSP26-A PE=2 SV=1 | 26.335 | 2.141 | 1.02394 × 10−3 | 1.098284796 | Up | 2.0505 | 8.3503 × 10−25 | Up | Up-Up |
| Cluster-1252.29574_orf1 | Unknown | 56.855 | 1.302 | 1.41228 × 10−2 | 0.380729449 | Up | 1.0417 | 3.2239 × 10−4 | Up | Up-Up |
| Cluster-1252.33343_orf1 | “Ornithine aminotransferase, mitochondrial OS=Arabidopsis thaliana GN=DELTA-OAT PE=1 SV=1” | 51.439 | 1.503 | 4.2365 × 10−8 | 0.587845009 | Up | 1.4006 | 2.5266 × 10−19 | Up | Up-Up |
| Cluster-1252.35376_orf1 | Unknown | 22.309 | 3.216 | 3.69926 × 10−13 | 1.685267407 | Up | 2.8743 | 1.7054 × 10−62 | Up | Up-Up |
| Cluster-1252.36163_orf1 | Expansin-like B1 OS=Arabidopsis thaliana GN=EXLB1 PE=2 SV=2 | 30.779 | 2.387 | 1.83814 × 10−2 | 1.255198566 | Up | 1.3227 | 1.0834 × 10−4 | Up | Up-Up |
| Cluster-1252.38004_orf2 | Protein C2-DOMAIN ABA-RELATED 9 OS=Arabidopsis thaliana GN=CAR9 PE=2 SV=1 | 21.642 | 1.534 | 7.6395 × 10−5 | 0.617298483 | Up | 1.4498 | 1.0054 × 10−10 | Up | Up-Up |
| Cluster-1252.39652_orf1 | 18 kDa seed maturation protein OS=Glycine max GN=GMPM1 PE=2 SV=1 | 12.611 | 1.805 | 1.62154 × 10−3 | 0.851998837 | Up | 2.2232 | 4.4188 × 10−12 | Up | Up-Up |
| Cluster-1252.41663_orf1 | Stress-related protein OS=Phaseolus vulgaris GN=SRP PE=2 SV=1 | 146.66 | 1.7 | 3.5979 × 10−5 | 0.765534746 | Up | 1.1772 | 1.5419 × 10−12 | Up | Up-Up |
| Cluster-1252.41945_orf1 | Glucose-1-phosphate adenylyltransferase large subunit 1 (Fragment) OS=Solanum tuberosum GN=AGPS1 PE=2 SV=1 | 59.046 | 2.277 | 4.6658 × 10−2 | 1.187134291 | Up | 2.2953 | 1.4116 × 10−4 | Up | Up-Up |
| Cluster-1252.42642_orf1 | Unknown | 16.163 | 3.21 | 9.99201 × 10−16 | 1.682573297 | Up | 1.6109 | 5.0453 × 10−27 | Up | Up-Up |
| Cluster-1252.42960_orf1 | Early nodulin-like protein 2 OS=Arabidopsis thaliana GN=At4g27520 PE=1 SV=1 | 103.53 | 1.698 | 1.90671 × 10−5 | 0.763836459 | Up | 1.0147 | 2.9677 × 10−13 | Up | Up-Up |
| Cluster-1252.42962_orf1 | Aldo-keto reductase family 4 member C9 OS=Arabidopsis thaliana GN=AKR4C9 PE=1 SV=1 | 36.025 | 1.447 | 5.56521 × 10−12 | 0.533064922 | Up | 1.5685 | 3.609 × 10−25 | Up | Up-Up |
| Cluster-1252.43381_orf1 | Thaumatin-like protein OS=Oryza sativa subsp. japonica GN=Os12g0628600 PE=1 SV=1 | 20.658 | 4.005 | 2.2805 × 10−3 | 2.001802243 | Up | 1.923 | 5.4445 × 10−14 | Up | Up-Up |
| Cluster-1252.43664_orf1 | Cinnamoyl-CoA reductase 1 OS=Arabidopsis thaliana GN=CCR1 PE=1 SV=1 | 34.902 | 2.775 | 1.0 × 10−32 | 1.472487771 | Up | 1.4878 | 7.6623 × 10−82 | Up | Up-Up |
| Cluster-1252.43700_orf1 | Vacuolar-processing enzyme OS=Vicia sativa PE=1 SV=1 | 148.25 | 1.998 | 2.2366 × 10−3 | 0.998556583 | Up | 1.237 | 4.6915 × 10−108 | Up | Up-Up |
| Cluster-1252.44442_orf1 | “Superoxide dismutase [Fe] 2, chloroplastic OS=Arabidopsis thaliana GN=FSD2 PE=1 SV=1” | 36.823 | 1.725 | 3.0282 × 10−10 | 0.786596362 | Up | 1.0925 | 1.076 × 10−3 | Up | Up-Up |
| Cluster-1252.45489_orf1 | Unknown | 87.568 | 1.436 | 2.8214 × 10−4 | 0.522055749 | Up | 1.1902 | 4.0089 × 10−11 | Up | Up-Up |
| Cluster-1252.45907_orf1 | Methylecgonone reductase OS=Erythroxylum coca PE=1 SV=1 | 35.219 | 1.928 | 1.2051 × 10−7 | 0.947105052 | Up | 1.071 | 1.6158 × 10−5 | Up | Up-Up |
| Cluster-1252.48706_orf1 | Delta-1-pyrroline-5-carboxylate synthase OS=Oryza sativa subsp. japonica GN=P5CS PE=2 SV=2 | 82.353 | 2.766 | 4.1957 × 10−8 | 1.467801156 | Up | 4.8411 | 2.0236 × 10−298 | Up | Up-Up |
| Cluster-1252.50314_orf1 | 1-aminocyclopropane-1-carboxylate oxidase homolog 5 OS=Arabidopsis thaliana GN=2A6 PE=2 SV=2 | 47.587 | 2.108 | 7.0431 × 10−11 | 1.075874867 | Up | 2.6988 | 1.3474 × 10−28 | Up | Up-Up |
| Cluster-1252.53143_orf1 | “1,4-alpha-glucan-branching enzyme 1, chloroplastic/amyloplastic (Fragment) OS=Pisum sativum GN=SBEII PE=1 SV=1” | 98.938 | 2.033 | 2.9402 × 10−7 | 1.023610215 | Up | 1.8785 | 8.7896 × 10−8 | Up | Up-Up |
| Cluster-1252.59078_orf1 | Pathogenesis-related protein PR-4B OS=Nicotiana tabacum PE=2 SV=1 | 19.406 | 2.258 | 3.0823 × 10−2 | 1.175045486 | Up | 2.4968 | 4.4984 × 10−5 | Up | Up-Up |
| Cluster-1252.62468_orf1 | Unknown | 107.76 | 8.756 | 6.3273 × 10−5 | 3.130271955 | Up | 3.225 | 2.9758 × 10−39 | Up | Up-Up |
| Cluster-1252.65950_orf1 | Galactinol--sucrose galactosyltransferase OS=Pisum sativum GN=RFS PE=1 SV=1 | 90.573 | 1.834 | 6.6447 × 10−10 | 0.874993639 | Up | 2.8804 | 2.2919 × 10−15 | Up | Up-Up |
| Cluster-1252.35865_orf1 | Tubulin alpha chain OS=Prunus dulcis GN=TUBA PE=2 SV=1 | 54.36 | 0.648 | 2.3014 × 10−3 | −0.625934282 | Down | −1.5125 | 1.0164 × 10−18 | Down | Down-Down |
| Cluster-1252.40636_orf1 | 1-aminocyclopropane-1-carboxylate oxidase OS=Prunus mume GN=ACO1 PE=2 SV=1 | 39.544 | 0.435 | 4.7576 × 10−6 | −1.200912694 | Down | −1.535 | 1.4095 × 10−47 | Down | Down-Down |
| Cluster-1252.43303_orf1 | “Ferredoxin--nitrite reductase, chloroplastic OS=Betula pendula GN=NIR1 PE=2 SV=1” | 69.24 | 0.423 | 1.0 × 10−32 | −1.241270432 | Down | −2.8277 | 4.993 × 10−52 | Down | Down-Down |
| Cluster-1252.43480_orf1 | “Protochlorophyllide reductase, chloroplastic OS=Pisum sativum GN=3PCR PE=1 SV=1” | 43.178 | 0.645 | 1.4187 × 10−11 | −0.632628934 | Down | −1.0364 | 1.1526 × 10−50 | Down | Down-Down |
| Cluster-1252.43550_orf1 | β-galactosidase 1 OS=Arabidopsis thaliana GN=BGAL1 PE=2 SV=1 | 93.798 | 0.506 | 2.2202 × 10−5 | −0.98279071 | Down | −1.5947 | 5.4211 × 10−54 | Down | Down-Down |
| Cluster-1252.44318_orf1 | Nitrate reductase [NADH] OS=Lotus japonicus GN=NIA PE=3 SV=1 | 102.62 | 0.623 | 1.33524 × 10−3 | −0.682695932 | Down | −2.0582 | 3.1915 × 10−25 | Down | Down-Down |
| Cluster-1252.45309_orf1 | Universal stress protein A-like protein OS=Arabidopsis thaliana GN=At3g01520 PE=1 SV=2 | 18.198 | 0.552 | 2.9101 × 10−2 | −0.857259828 | Down | −1.2288 | 1.3773 × 10−29 | Down | Down-Down |
| Cluster-1252.48945_orf1 | Unknown | 40.668 | 0.512 | 2.36871 × 10−11 | −0.965784285 | Down | −2.4873 | 1.4274 × 10−10 | Down | Down-Down |
| Protein Accession | Protein Description | MW (kDa) | M25W/M25CK Ratio | M25W/M25CK p-Value | Protein LOG2 M25W/M25CK | Regulation | Transcription LOG2 M25W/M25CK | p adj | Regulation | Type |
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster-1252.18690_orf1 | Unknown | 42.266 | 1.345 | 4.7545 × 10−2 | 0.427606173 | Up | 2.7094 | 1.029 × 10−14 | Up | Up-Up |
| Cluster-1252.25469_orf1 | Probable glutathione S-transferase OS=Glycine max GN=HSP26-A PE=2 SV=1 | 26.335 | 2.541 | 1.12492 × 10−3 | 1.345396375 | Up | 1.7162 | 7.0349 × 10−27 | Up | Up-Up |
| Cluster-1252.27678_orf1 | Cinnamoyl-CoA reductase 2 OS=Arabidopsis thaliana GN=CCR2 PE=1 SV=1 | 37.481 | 1.36 | 3.5464 × 10−3 | 0.443606651 | Up | 3.4894 | 1.0739 × 10−7 | Up | Up-Up |
| Cluster-1252.32933_orf1 | Probable cinnamyl alcohol dehydrogenase OS=Medicago sativa GN=CAD2 PE=1 SV=1 | 40.598 | 1.336 | 3.82 × 10−4 | 0.417920008 | Up | 1.0817 | 2.6648 × 10−7 | Up | Up-Up |
| Cluster-1252.33343_orf1 | “Ornithine aminotransferase, mitochondrial OS=Arabidopsis thaliana GN=DELTA-OAT PE=1 SV=1” | 51.439 | 1.813 | 3.3792 × 10−8 | 0.858378925 | Up | 2.6159 | 1.0261 × 10−81 | Up | Up-Up |
| Cluster-1252.35478_orf1 | Unknown | 46.282 | 1.401 | 1.0601 × 10−9 | 0.486456956 | Up | 1.59 | 1.3747 × 10−13 | Up | Up-Up |
| Cluster-1252.36163_orf1 | Expansin-like B1 OS=Arabidopsis thaliana GN=EXLB1 PE=2 SV=2 | 30.779 | 2.054 | 1.2356 × 10−2 | 1.038436182 | Up | 2.62 | 1.3061 × 10−10 | Up | Up-Up |
| Cluster-1252.36198_orf1 | Cytochrome b5 OS=Brassica oleracea var. botrytis GN=CYB5 PE=1 SV=1 | 17.483 | 1.312 | 1.59783 × 10−4 | 0.39176772 | Up | 1.1567 | 2.1863 × 10−10 | Up | Up-Up |
| Cluster-1252.38004_orf2 | Protein C2-DOMAIN ABA-RELATED 9 OS=Arabidopsis thaliana GN=CAR9 PE=2 SV=1 | 21.642 | 1.651 | 6.0141 × 10−5 | 0.72334012 | Up | 1.2091 | 4.96 × 10−7 | Up | Up-Up |
| Cluster-1252.39652_orf1 | 18 kDa seed maturation protein OS=Glycine max GN=GMPM1 PE=2 SV=1 | 12.611 | 2.227 | 4.7834 × 10−3 | 1.155101558 | Up | 1.393 | 1.1811 × 10−4 | Up | Up-Up |
| Cluster-1252.42092_orf1 | Desiccation protectant protein Lea14 homolog OS=Glycine max PE=2 SV=1 | 51.585 | 1.35 | 1.00421 × 10−3 | 0.432959407 | Up | 1.8427 | 4.6211 × 10−8 | Up | Up-Up |
| Cluster-1252.42169_orf1 | 4-hydroxyphenylpyruvate dioxygenase OS=Arabidopsis thaliana GN=HPD PE=1 SV=2 | 59.596 | 1.838 | 3.7081 × 10−5 | 0.878136767 | Up | 2.1711 | 3.1841 × 10−81 | Up | Up-Up |
| Cluster-1252.42520_orf1 | ABC transporter C family member 4 OS=Arabidopsis thaliana GN=ABCC4 PE=2 SV=2 | 194.56 | 1.426 | 1.39367 × 10−2 | 0.511973982 | Up | 1.1124 | 7.8224 × 10−50 | Up | Up-Up |
| Cluster-1252.42642_orf1 | Unknown | 16.163 | 2.542 | 1.56475 × 10−12 | 1.34596403 | Up | 2.1592 | 1.2647 × 10−37 | Up | Up-Up |
| Cluster-1252.42962_orf1 | Aldo-keto reductase family 4 member C9 OS=Arabidopsis thaliana GN=AKR4C9 PE=1 SV=1 | 36.025 | 1.711 | 1.11022 × 10−16 | 0.77483976 | Up | 1.6703 | 1.2377 × 10−41 | Up | Up-Up |
| Cluster-1252.43664_orf1 | Cinnamoyl-CoA reductase 1 OS=Arabidopsis thaliana GN=CCR1 PE=1 SV=1 | 34.902 | 2.885 | 1.0 × 10−32 | 1.528571319 | Up | 1.0253 | 4.2827 × 10−31 | Up | Up-Up |
| Cluster-1252.43700_orf1 | Vacuolar-processing enzyme OS=Vicia sativa PE=1 SV=1 | 148.25 | 2.293 | 7.1779 × 10−4 | 1.197236355 | Up | 1.497 | 1.8971 × 10−177 | Up | Up-Up |
| Cluster-1252.43923_orf1 | “Crocetin glucosyltransferase, chloroplastic OS=Gardenia jasminoides GN=UGT75L6 PE=1 SV=1” | 54.447 | 1.499 | 1.80426 × 10−4 | 0.584000383 | Up | 1.3321 | 1.6138 × 10−5 | Up | Up-Up |
| Cluster-1252.44444_orf1 | Malonate-CoA ligase OS=Arabidopsis thaliana GN=AAE13 PE=1 SV=1 | 69.952 | 1.312 | 1.37048 × 10−4 | 0.39176772 | Up | 1.9792 | 1.2674 × 10−22 | Up | Up-Up |
| Cluster-1252.45456_orf1 | Copper transport protein CCH OS=Arabidopsis thaliana GN=CCH PE=1 SV=1 | 15.409 | 2.06 | 2.3795 × 10−2 | 1.042644337 | Up | 1.0154 | 4.6715 × 10−3 | Up | Up-Up |
| Cluster-1252.45489_orf1 | Unknown | 87.568 | 1.487 | 9.5623 × 10−4 | 0.572404647 | Up | 1.0995 | 7.6879 × 10−12 | Up | Up-Up |
| Cluster-1252.46922_orf1 | Aldehyde dehydrogenase family 7 member A1 OS=Pisum sativum PE=1 SV=3 | 54.83 | 1.407 | 4.1199 × 10−7 | 0.492622329 | Up | 2.4112 | 1.3416 × 10−169 | Up | Up-Up |
| Cluster-1252.47893_orf1 | Unknown | 42.991 | 1.503 | 7.1367 × 10−3 | 0.587845009 | Up | 2.4434 | 1.0982 × 10−38 | Up | Up-Up |
| Cluster-1252.48706_orf1 | Delta-1-pyrroline-5-carboxylate synthase OS=Oryza sativa subsp. japonica GN=P5CS PE=2 SV=2 | 82.353 | 2.771 | 2.0696 × 10−7 | 1.470406711 | Up | 6.2581 | 7.2562 × 10−296 | Up | Up-Up |
| Cluster-1252.49757_orf2 | Unknown | 28.735 | 1.329 | 1.89241 × 10−2 | 0.410341105 | Up | 1.6066 | 2.6525 × 10−10 | Up | Up-Up |
| Cluster-1252.49800_orf1 | β-galactosidase 8 OS=Arabidopsis thaliana GN=BGAL8 PE=2 SV=2 | 65.645 | 1.349 | 1.14055 × 10−3 | 0.431890348 | Up | 4.143 | 1.8906 × 10−80 | Up | Up-Up |
| Cluster-1252.50314_orf1 | 1-aminocyclopropane-1-carboxylate oxidase homolog 5 OS=Arabidopsis thaliana GN=2A6 PE=2 SV=2 | 47.587 | 2.728 | 1.66533 × 10−15 | 1.447843644 | Up | 3.2213 | 3.1324 × 10−40 | Up | Up-Up |
| Cluster-1252.53143_orf1 | “1,4-α-glucan-branching enzyme 1, chloroplastic/amyloplastic (Fragment) OS=Pisum sativum GN=SBEII PE=1 SV=1” | 98.938 | 1.681 | 3.2469 × 10−6 | 0.749319725 | Up | 1.6874 | 7.8729 × 10−7 | Up | Up-Up |
| Cluster-1252.62468_orf1 | Unknown | 107.76 | 6.211 | 2.2964 × 10−5 | 2.634825568 | Up | 4.7334 | 5.8563 × 10−42 | Up | Up-Up |
| Cluster-1252.65950_orf1 | Galactinol-sucrose galactosyltransferase OS=Pisum sativum GN=RFS PE=1 SV=1 | 90.573 | 1.73 | 1.9153 × 10−10 | 0.790772038 | Up | 2.7024 | 3.5766 × 10−5 | Up | Up-Up |
| Cluster-1252.38137_orf1 | “Superoxide dismutase [Cu-Zn], chloroplastic OS=Medicago sativa GN=SODCP PE=2 SV=1” | 24.007 | 0.76 | 3.2114 × 10−4 | −0.395928676 | Down | −3.946 | 5.8488 × 10−29 | Down | Down-Down |
| Cluster-1252.40018_orf1 | “CBS domain-containing protein CBSX3, mitochondrial OS=Arabidopsis thaliana GN=CBSX3 PE=1 SV=1” | 26.605 | 0.752 | 1.1475 × 10−11 | −0.411195433 | Down | −1.274 | 3.2125 × 10−18 | Down | Down-Down |
| Cluster-1252.40357_orf1 | Unknown | 23.476 | 0.658 | 9.9921 × 10−4 | −0.603840511 | Down | −1.2781 | 3.4128 × 10−4 | Down | Down-Down |
| Cluster-1252.40636_orf1 | 1-aminocyclopropane-1-carboxylate oxidase OS=Prunus mume GN=ACO1 PE=2 SV=1 | 39.544 | 0.544 | 3.9251 × 10−5 | −0.878321443 | Down | −1.0541 | 5.7398 × 10−15 | Down | Down-Down |
| Cluster-1252.42669_orf1 | “50S ribosomal protein L19, chloroplastic OS=Spinacia oleracea GN=RPL19 PE=1 SV=2” | 27.445 | 0.737 | 1.39429 × 10−2 | −0.440263476 | Down | −1.1371 | 9.5976 × 10−9 | Down | Down-Down |
| Cluster-1252.43155_orf1 | “30S ribosomal protein S17, chloroplastic (Fragment) OS=Pisum sativum GN=RPS17 PE=2 SV=1” | 19.162 | 0.764 | 4.0527 × 10−5 | −0.388355457 | Down | −1.3192 | 2.386 × 10−7 | Down | Down-Down |
| Cluster-1252.43183_orf1 | Protein TSS OS=Arabidopsis thaliana GN=TSS PE=1 SV=1 | 180.86 | 0.718 | 4.32987 × 10−15 | −0.477944251 | Down | −1.2443 | 3.8189 × 10−29 | Down | Down-Down |
| Cluster-1252.43187_orf1 | “Thiamine thiazole synthase 2, chloroplastic OS=Vitis vinifera GN=THI1-2 PE=3 SV=1” | 41.382 | 0.63 | 1.0979 × 10−7 | −0.666576266 | Down | −1.5012 | 3.85 × 10−204 | Down | Down-Down |
| Cluster-1252.43232_orf1 | “Probable carotenoid cleavage dioxygenase 4, chloroplastic OS=Arabidopsis thaliana GN=CCD4 PE=1 SV=1” | 40.321 | 0.705 | 5.9396 × 10−8 | −0.504304837 | Down | −1.5008 | 4.5918 × 10−20 | Down | Down-Down |
| Cluster-1252.43303_orf1 | “Ferredoxin--nitrite reductase, chloroplastic OS=Betula pendula GN=NIR1 PE=2 SV=1” | 69.24 | 0.482 | 1.0 × 10−32 | −1.052894948 | Down | −1.6604 | 1.2831 × 10−26 | Down | Down-Down |
| Cluster-1252.43381_orf1 | Thaumatin-like protein OS=Oryza sativa subsp. japonica GN=Os12g0628600 PE=1 SV=1 | 20.658 | 0.707 | 3.3377 × 10−2 | −0.50021788 | Down | −2.7087 | 1.6235 × 10−50 | Down | Down-Down |
| Cluster-1252.43480_orf1 | “Protochlorophyllide reductase, chloroplastic OS=Pisum sativum GN=3PCR PE=1 SV=1” | 43.178 | 0.503 | 4.6423 × 10−9 | −0.991369695 | Down | −1.7186 | 7.5419 × 10−93 | Down | Down-Down |
| Cluster-1252.43489_orf1 | “Ketol-acid reductoisomerase, chloroplastic OS=Pisum sativum GN=PGAAIR PE=2 SV=1” | 66.112 | 0.679 | 1.0 × 10−32 | −0.55851652 | Down | −1.0004 | 3.72 × 10−8 | Down | Down-Down |
| Cluster-1252.43517_orf1 | “Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplastic OS=Euphorbia esula GN=CRD1 PE=3 SV=1” | 48.801 | 0.633 | 1.9456 × 10−10 | −0.659722595 | Down | −1.5422 | 1.5614 × 10−85 | Down | Down-Down |
| Cluster-1252.43550_orf1 | β-galactosidase 1 OS=Arabidopsis thaliana GN=BGAL1 PE=2 SV=1 | 93.798 | 0.442 | 2.3143 × 10−5 | −1.177881725 | Down | −1.9732 | 1.0767 × 10−53 | Down | Down-Down |
| Cluster-1252.43873_orf1 | “Light-harvesting complex-like protein 3 isotype 1, chloroplastic OS=Arabidopsis thaliana GN=LIL3.1 PE=1 SV=1” | 29.313 | 0.761 | 1.4038 × 10−6 | −0.394031641 | Down | −1.6761 | 3.8258 × 10−18 | Down | Down-Down |
| Cluster-1252.43934_orf1 | “Probable carotenoid cleavage dioxygenase 4, chloroplastic OS=Arabidopsis thaliana GN=CCD4 PE=1 SV=1” | 27.699 | 0.746 | 1.28296 × 10−3 | −0.422752464 | Down | −1.6188 | 2.6379 × 10−10 | Down | Down-Down |
| Cluster-1252.44318_orf1 | Nitrate reductase [NADH] OS=Lotus japonicus GN=NIA PE=3 SV=1 | 102.62 | 0.573 | 2.1975 × 10−4 | −0.803392956 | Down | −1.7715 | 2.6347 × 10−42 | Down | Down-Down |
| Cluster-1252.45201_orf2 | “Magnesium-chelatase subunit ChlI, chloroplastic OS=Glycine max GN=CHLI PE=2 SV=1” | 47.591 | 0.681 | 1.8359 × 10−11 | −0.554273297 | Down | −1.8322 | 5.4646 × 10−38 | Down | Down-Down |
| Cluster-1252.45508_orf1 | “Porphobilinogen deaminase, chloroplastic OS=Pisum sativum GN=HEMC PE=1 SV=1” | 45.052 | 0.665 | 1.0314 × 10−12 | −0.588573754 | Down | −1.0198 | 2.5422 × 10−3 | Down | Down-Down |
| Cluster-1252.46447_orf1 | “Magnesium protoporphyrin IX methyltransferase, chloroplastic OS=Arabidopsis thaliana GN=CHLM PE=1 SV=1” | 39.446 | 0.724 | 2.9589 × 10−4 | −0.465938398 | Down | −3.5109 | 3.9122 × 10−25 | Down | Down-Down |
| Cluster-1252.47724_orf1 | Stem 28 kDa glycoprotein OS=Glycine max GN=VSPA PE=2 SV=1 | 30.865 | 0.753 | 1.58697 × 10−5 | −0.40927823 | Down | −1.5366 | 4.9784 × 10−7 | Down | Down-Down |
| Cluster-1252.48703_orf1 | “Glutamyl-tRNA reductase 1, chloroplastic OS=Cucumis sativus GN=HEMA1 PE=2 SV=1” | 59.248 | 0.541 | 5.1604 × 10−3 | −0.886299501 | Down | −1.1181 | 2.0974 × 10−5 | Down | Down-Down |
| Cluster-1252.48945_orf1 | Unknown | 40.668 | 0.611 | 7.1715 × 10−11 | −0.710755715 | Down | −3.236 | 6.0036 × 10−22 | Down | Down-Down |
| Cluster-1252.49560_orf1 | “Probable plastid-lipid-associated protein 8, chloroplastic OS=Arabidopsis thaliana GN=PAP8 PE=1 SV=1” | 168.8 | 0.755 | 3.6428 × 10−4 | −0.40545145 | Down | −1.072 | 1.7811 × 10−6 | Down | Down-Down |
| Cluster-1252.51383_orf1 | “50S ribosomal protein L29, chloroplastic OS=Arabidopsis thaliana GN=RPL29 PE=1 SV=1” | 20 | 0.756 | 1.67585 × 10−3 | −0.40354186 | Down | −1.6621 | 2.8565 × 10−8 | Down | Down-Down |
| Cluster-1252.59847_orf1 | ATP sulfurylase 2 OS=Arabidopsis thaliana GN=APS2 PE=1 SV=1 | 57.663 | 0.633 | 1.8719 × 10−7 | −0.659722595 | Down | −1.5753 | 1.7315 × 10−3 | Down | Down-Down |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, N.; Yang, Z.; Zhang, Z.; Hu, L.; Chen, L. Comparative Transcriptome Combined with Proteome Analyses Revealed Key Factors Involved in Alfalfa (Medicago sativa) Response to Waterlogging Stress. Int. J. Mol. Sci. 2019, 20, 1359. https://doi.org/10.3390/ijms20061359
Zeng N, Yang Z, Zhang Z, Hu L, Chen L. Comparative Transcriptome Combined with Proteome Analyses Revealed Key Factors Involved in Alfalfa (Medicago sativa) Response to Waterlogging Stress. International Journal of Molecular Sciences. 2019; 20(6):1359. https://doi.org/10.3390/ijms20061359
Chicago/Turabian StyleZeng, Ningbo, Zhijian Yang, Zhifei Zhang, Longxing Hu, and Liang Chen. 2019. "Comparative Transcriptome Combined with Proteome Analyses Revealed Key Factors Involved in Alfalfa (Medicago sativa) Response to Waterlogging Stress" International Journal of Molecular Sciences 20, no. 6: 1359. https://doi.org/10.3390/ijms20061359
APA StyleZeng, N., Yang, Z., Zhang, Z., Hu, L., & Chen, L. (2019). Comparative Transcriptome Combined with Proteome Analyses Revealed Key Factors Involved in Alfalfa (Medicago sativa) Response to Waterlogging Stress. International Journal of Molecular Sciences, 20(6), 1359. https://doi.org/10.3390/ijms20061359







