Apoptosis Induction byHistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70
Abstract
:1. Introduction
2. Classification of HDACs and Cellular Location
3. Biological Functions of HDACs
4. HDACi for Cancer Therapy
5. Apoptosis Induction by HDACis
6. Ku70 Regulates Apoptosis-Related Proteins
6.1. Bax
6.2. Mcl-1
6.3. c-FLIP
7. Ku70 Acetylation by HDACi Treatment
8. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Bezecny, P. Histone deacetylase inhibitors in glioblastoma: Pre-clinical and clinical experience. Med. Oncol. 2014, 31, 985. [Google Scholar] [CrossRef]
- Krusche, C.A.; Wulfing, P.; Kersting, C.; Vloet, A.; Bocker, W.; Kiesel, L.; Beier, H.M.; Alfer, J. Histone deacetylase-1 and -3 protein expression in human breast cancer: A tissue microarray analysis. Breast Cancer Res. Treat. 2005, 90, 15–23. [Google Scholar] [CrossRef]
- Minamiya, Y.; Ono, T.; Saito, H.; Takahashi, N.; Ito, M.; Mitsui, M.; Motoyama, S.; Ogawa, J. Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer 2011, 74, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Rikimaru, T.; Taketomi, A.; Yamashita, Y.; Shirabe, K.; Hamatsu, T.; Shimada, M.; Maehara, Y. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology 2007, 72, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Roske, A.; Gekeler, V.; Beckers, T.; Ebert, M.P.; Pross, M.; Dietel, M.; Denkert, C.; Rocken, C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008, 9, 139–148. [Google Scholar] [CrossRef]
- Weichert, W.; Roske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Fritzsche, F.R.; Niesporek, S.; Denkert, C.; Dietel, M.; et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer 2008, 98, 604–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weichert, W.; Roske, A.; Niesporek, S.; Noske, A.; Buckendahl, A.C.; Dietel, M.; Gekeler, V.; Boehm, M.; Beckers, T.; Denkert, C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: Specific role of class I histone deacetylases in vitro and in vivo. Clin. Cancer Res. 2008, 14, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Niegisch, G.; Knievel, J.; Koch, A.; Hader, C.; Fischer, U.; Albers, P.; Schulz, W.A. Changes in histone deacetylase (HDAC) expression patterns and activity of HDAC inhibitors in urothelial cancers. Urol. Oncol. 2013, 31, 1770–1779. [Google Scholar] [CrossRef]
- West, A.C.; Johnstone, R.W. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Investig. 2014, 124, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Bishayee, A.; Pandey, A.K. Targeting Histone Deacetylases with Natural and Synthetic Agents: An Emerging Anticancer Strategy. Nutrients 2018, 10, 731. [Google Scholar] [CrossRef]
- Suraweera, A.; O’Byrne, K.J.; Richard, D.J. Combination Therapy with Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 2018, 8, 92. [Google Scholar] [CrossRef]
- Wawruszak, A.; Kalafut, J.; Okon, E.; Czapinski, J.; Halasa, M.; Przybyszewska, A.; Miziak, P.; Okla, K.; Rivero-Muller, A.; Stepulak, A. Histone Deacetylase Inhibitors and Phenotypical Transformation of Cancer Cells. Cancers 2019, 11, 148. [Google Scholar] [CrossRef]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef]
- Yang, X.J.; Seto, E. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef]
- Yoo, C.B.; Jones, P.A. Epigenetic therapy of cancer: Past, present and future. Nat. Rev. Drug Discov. 2006, 5, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.Y.; Lavu, S.; Bitterman, K.J.; Hekking, B.; Imahiyerobo, T.A.; Miller, C.; Frye, R.; Ploegh, H.; Kessler, B.M.; Sinclair, D.A. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell 2004, 13, 627–638. [Google Scholar] [CrossRef]
- Costanzo, A.; Merlo, P.; Pediconi, N.; Fulco, M.; Sartorelli, V.; Cole, P.A.; Fontemaggi, G.; Fanciulli, M.; Schiltz, L.; Blandino, G.; et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell 2002, 9, 175–186. [Google Scholar] [CrossRef]
- Huang, B.H.; Laban, M.; Leung, C.H.; Lee, L.; Lee, C.K.; Salto-Tellez, M.; Raju, G.C.; Hooi, S.C. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005, 12, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Olaharski, A.J.; Rine, J.; Marshall, B.L.; Babiarz, J.; Zhang, L.; Verdin, E.; Smith, M.T. The flavoring agent dihydrocoumarin reverses epigenetic silencing and inhibits sirtuin deacetylases. PLoS Genet. 2005, 1, e77. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Kwok, R.P. Regulation of ku70-bax complex in cells. J. Cell Death 2014, 7, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef]
- Groselj, B.; Sharma, N.L.; Hamdy, F.C.; Kerr, M.; Kiltie, A.E. Histone deacetylase inhibitors as radiosensitisers: Effects on DNA damage signalling and repair. Br. J. Cancer 2013, 108, 748–754. [Google Scholar] [CrossRef]
- Blander, G.; Guarente, L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004, 73, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Denu, J.M. Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta 2009, 1789, 45–57. [Google Scholar] [CrossRef]
- Glass, C.K.; Rosenfeld, M.G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000, 14, 121–141. [Google Scholar]
- Garmpis, N.; Damaskos, C.; Garmpi, A.; Dimitroulis, D.; Spartalis, E.; Margonis, G.A.; Schizas, D.; Deskou, I.; Doula, C.; Magkouti, E.; et al. Targeting Histone Deacetylases in Malignant Melanoma: A Future Therapeutic Agent or Just Great Expectations? Anticancer Res. 2017, 37, 5355–5362. [Google Scholar] [PubMed]
- Singh, B.N.; Zhang, G.; Hwa, Y.L.; Li, J.; Dowdy, S.C.; Jiang, S.W. Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer Ther. 2010, 10, 935–954. [Google Scholar] [CrossRef] [Green Version]
- Ito, A.; Lai, C.H.; Zhao, X.; Saito, S.; Hamilton, M.H.; Appella, E.; Yao, T.P. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001, 20, 1331–1340. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Zhao, W.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation is indispensable for p53 activation. Cell 2008, 133, 612–626. [Google Scholar] [CrossRef]
- Brooks, C.L.; Gu, W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2011, 2, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Su, F.; Chen, D.; Shiloh, A.; Gu, W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 2000, 408, 377–381. [Google Scholar] [CrossRef]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001, 107, 137–148. [Google Scholar] [CrossRef]
- Vaziri, H.; Dessain, S.K.; Ng Eaton, E.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef]
- Ding, G.; Liu, H.D.; Huang, Q.; Liang, H.X.; Ding, Z.H.; Liao, Z.J.; Huang, G. HDAC6 promotes hepatocellular carcinoma progression by inhibiting P53 transcriptional activity. FEBS Lett. 2013, 587, 880–886. [Google Scholar] [CrossRef] [Green Version]
- Mrakovcic, M.; Bohner, L.; Hanisch, M.; Frohlich, L.F. Epigenetic Targeting of Autophagy via HDAC Inhibition in Tumor Cells: Role of p53. Int. J. Mol. Sci. 2018, 19, 3952. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.E.; Ghosh, G. Regulation of DNA binding by Rel/NF-kappaB transcription factors: Structural views. Oncogene 1999, 18, 6845–6852. [Google Scholar] [CrossRef]
- Ito, K. Impact of post-translational modifications of proteins on the inflammatory process. Biochem. Soc. Trans. 2007, 35, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fischle, W.; Verdin, E.; Greene, W.C. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 2001, 293, 1653–1657. [Google Scholar] [CrossRef]
- Ashburner, B.P.; Westerheide, S.D.; Baldwin, A.S., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 2001, 21, 7065–7077. [Google Scholar] [CrossRef] [PubMed]
- Imre, G.; Gekeler, V.; Leja, A.; Beckers, T.; Boehm, M. Histone deacetylase inhibitors suppress the inducibility of nuclear factor-kappaB by tumor necrosis factor-alpha receptor-1 down-regulation. Cancer Res. 2006, 66, 5409–5418. [Google Scholar] [CrossRef]
- Takada, Y.; Gillenwater, A.; Ichikawa, H.; Aggarwal, B.B. Suberoylanilide hydroxamic acid potentiates apoptosis, inhibits invasion, and abolishes osteoclastogenesis by suppressing nuclear factor-kappaB activation. J. Biol. Chem. 2006, 281, 5612–5622. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, J.S.; Xu, W.; Neckers, L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell 2003, 3, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Nimmanapalli, R.; O’Bryan, E.; Bhalla, K. Geldanamycin and its analogue 17-allylamino-17-demethoxygeldanamycin lowers Bcr-Abl levels and induces apoptosis and differentiation of Bcr-Abl-positive human leukemic blasts. Cancer Res. 2001, 61, 1799–1804. [Google Scholar]
- George, P.; Bali, P.; Cohen, P.; Tao, J.; Guo, F.; Sigua, C.; Vishvanath, A.; Fiskus, W.; Scuto, A.; Annavarapu, S.; et al. Cotreatment with 17-allylamino-demethoxygeldanamycin and FLT-3 kinase inhibitor PKC412 is highly effective against human acute myelogenous leukemia cells with mutant FLT-3. Cancer Res. 2004, 64, 3645–3652. [Google Scholar] [CrossRef] [PubMed]
- Bagatell, R.; Whitesell, L. Altered Hsp90 function in cancer: A unique therapeutic opportunity. Mol. Cancer Ther. 2004, 3, 1021–1030. [Google Scholar]
- Kramer, O.H.; Mahboobi, S.; Sellmer, A. Drugging the HDAC6-HSP90 interplay in malignant cells. Trends Pharmacol. Sci. 2014, 35, 501–509. [Google Scholar] [CrossRef] [PubMed]
- George, P.; Bali, P.; Annavarapu, S.; Scuto, A.; Fiskus, W.; Guo, F.; Sigua, C.; Sondarva, G.; Moscinski, L.; Atadja, P.; et al. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood 2005, 105, 1768–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, K.; Fiskus, W.; Choi, D.S.; Bhaskara, S.; Cerchietti, L.; Devaraj, S.G.; Shah, B.; Sharma, S.; Chang, J.C.; Melnick, A.M.; et al. Histone deacetylase inhibitor treatment induces ‘BRCAness’ and synergistic lethality with PARP inhibitor and cisplatin against human triple negative breast cancer cells. Oncotarget 2014, 5, 5637–5650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, D.E.; Darnell, J.E., Jr. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Han, J.J.; Stenson, M.; Wellik, L.; Witzig, T.E. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: Implications for therapy. Leukemia 2012, 26, 1356–1364. [Google Scholar] [CrossRef]
- Sundaresan, N.R.; Pillai, V.B.; Wolfgeher, D.; Samant, S.; Vasudevan, P.; Parekh, V.; Raghuraman, H.; Cunningham, J.M.; Gupta, M.; Gupta, M.P. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 2011, 4, ra46. [Google Scholar] [CrossRef] [PubMed]
- Pillai, V.B.; Sundaresan, N.R.; Gupta, M.P. Regulation of Akt signaling by sirtuins: Its implication in cardiac hypertrophy and aging. Circ. Res. 2014, 114, 368–378. [Google Scholar] [CrossRef]
- Iaconelli, J.; Lalonde, J.; Watmuff, B.; Liu, B.; Mazitschek, R.; Haggarty, S.J.; Karmacharya, R. Lysine Deacetylation by HDAC6 Regulates the Kinase Activity of AKT in Human Neural Progenitor Cells. ACS Chem. Biol. 2017, 12, 2139–2148. [Google Scholar] [CrossRef]
- Long, J.; Fang, W.Y.; Chang, L.; Gao, W.H.; Shen, Y.; Jia, M.Y.; Zhang, Y.X.; Wang, Y.; Dou, H.B.; Zhang, W.J.; et al. Targeting HDAC3, a new partner protein of AKT in the reversal of chemoresistance in acute myeloid leukemia via DNA damage response. Leukemia 2017, 31, 2761–2770. [Google Scholar] [CrossRef] [PubMed]
- Kauppinen, A.; Suuronen, T.; Ojala, J.; Kaarniranta, K.; Salminen, A. Antagonistic crosstalk between NF-kappaB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell. Signal. 2013, 25, 1939–1948. [Google Scholar] [CrossRef]
- Rothgiesser, K.M.; Erener, S.; Waibel, S.; Luscher, B.; Hottiger, M.O. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci. 2010, 123, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ye, J.; Chen, X.; Shi, J.; Wu, W.; Lin, W.; Lin, W.; Li, Y.; Fu, H.; Li, S. Valproic acid attenuates traumatic spinal cord injury-induced inflammation via STAT1 and NF-kappaB pathway dependent of HDAC3. J. Neuroinflamm. 2018, 15, 150. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Li, K.; Li, Y.; Liu, D.; Zhao, L.; Jing, Y. HDAC and Ku70 axis- an effective target for apoptosis induction by a new 2-cyano-3-oxo-1,9-dien glycyrrhetinic acid analogue. Cell Death Dis. 2018, 9, 623. [Google Scholar] [CrossRef]
- Lee, S.J.; Jang, H.; Park, C. Maspin increases Ku70 acetylation and Bax-mediated cell death in cancer cells. Int. J. Mol. Med. 2012, 29, 225–230. [Google Scholar]
- Subramanian, C.; Jarzembowski, J.A.; Opipari, A.W., Jr.; Castle, V.P.; Kwok, R.P. HDAC6 deacetylates Ku70 and regulates Ku70-Bax binding in neuroblastoma. Neoplasia 2011, 13, 726–734. [Google Scholar] [CrossRef]
- Roth, M.; Wang, Z.; Chen, W.Y. SIRT1 and LSD1 competitively regulate KU70 functions in DNA repair and mutation acquisition in cancer cells. Oncotarget 2016, 7, 50195–50214. [Google Scholar] [CrossRef] [Green Version]
- Sundaresan, N.R.; Samant, S.A.; Pillai, V.B.; Rajamohan, S.B.; Gupta, M.P. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell. Biol. 2008, 28, 6384–6401. [Google Scholar] [CrossRef]
- Tao, N.N.; Ren, J.H.; Tang, H.; Ran, L.K.; Zhou, H.Z.; Liu, B.; Huang, A.L.; Chen, J. Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017, 485, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, A.; Shimazu, T.; Sumida, Y.; Saito, A.; Yoshimatsu, Y.; Seigneurin-Berny, D.; Osada, H.; Komatsu, Y.; Nishino, N.; Khochbin, S.; et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002, 21, 6820–6831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Zhuang, Q.; Li, W.; Benda, C.; Huang, Z.; Ahmed, T.; Liu, P.; Guo, X.; Ibanez, D.P.; Luo, Z.; Zhang, M.; et al. NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat. Cell Biol. 2018, 20, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X.; Polakiewicz, R.D.; Yao, T.P.; Comb, M.J. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J. Biol. Chem. 2008, 283, 12686–12690. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Sue, Y.M.; Yang, N.J.; Lee, Y.H.; Juan, S.H. 3-Methylcholanthrene, an AhR agonist, caused cell-cycle arrest by histone deacetylation through a RhoA-dependent recruitment of HDAC1 and pRb2 to E2F1 complex. PLoS ONE 2014, 9, e92793. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.W.; Bae, M.K.; Ahn, M.Y.; Kim, S.H.; Sohn, T.K.; Bae, M.H.; Yoo, M.A.; Song, E.J.; Lee, K.J.; Kim, K.W. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef]
- Yang, F.Q.; Liu, M.; Yang, F.P.; Che, J.; Li, W.; Zhai, W.; Wang, G.C.; Zheng, J.H.; Li, X. VPA inhibits renal cancer cell migration by targeting HDAC2 and down-regulating HIF-1alpha. Mol. Biol. Rep. 2014, 41, 1511–1518. [Google Scholar] [CrossRef]
- Geng, H.; Harvey, C.T.; Pittsenbarger, J.; Liu, Q.; Beer, T.M.; Xue, C.; Qian, D.Z. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J. Biol. Chem. 2011, 286, 38095–38102. [Google Scholar] [CrossRef]
- Ye, M.; Fang, Z.; Gu, H.; Song, R.; Ye, J.; Li, H.; Wu, Z.; Zhou, S.; Li, P.; Cai, X.; et al. Histone deacetylase 5 promotes the migration and invasion of hepatocellular carcinoma via increasing the transcription of hypoxia-inducible factor-1alpha under hypoxia condition. Tumour Biol. 2017, 39, 1010428317705034. [Google Scholar] [CrossRef] [PubMed]
- Kawai, H.; Li, H.; Avraham, S.; Jiang, S.; Avraham, H.K. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int. J. Cancer 2003, 107, 353–358. [Google Scholar] [CrossRef]
- Iaconelli, J.; Huang, J.H.; Berkovitch, S.S.; Chattopadhyay, S.; Mazitschek, R.; Schreiber, S.L.; Haggarty, S.J.; Karmacharya, R. HDAC6 inhibitors modulate Lys49 acetylation and membrane localization of beta-catenin in human iPSC-derived neuronal cells. ACS Chem. Biol. 2015, 10, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, Y.; Towatari, M.; Tsuzuki, S.; Hayakawa, F.; Maeda, T.; Miyata, Y.; Tanimoto, M.; Saito, H. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood 2001, 98, 2116–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watamoto, K.; Towatari, M.; Ozawa, Y.; Miyata, Y.; Okamoto, M.; Abe, A.; Naoe, T.; Saito, H. Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation. Oncogene 2003, 22, 9176–9184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Horst, A.; Tertoolen, L.G.; de Vries-Smits, L.M.; Frye, R.A.; Medema, R.H.; Burgering, B.M. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 2004, 279, 28873–28879. [Google Scholar] [CrossRef]
- Zhang, H.; Shang, Y.P.; Chen, H.Y.; Li, J. Histone deacetylases function as novel potential therapeutic targets for cancer. Hepatol. Res. 2017, 47, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncology 2007, 12, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.; Rahman, F.; Piekarz, R.; Peer, C.; Frye, R.; Robey, R.W.; Gardner, E.R.; Figg, W.D.; Bates, S.E. Romidepsin: A new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev. Anticancer Ther. 2010, 10, 997–1008. [Google Scholar] [CrossRef]
- Sawas, A.; Radeski, D.; O’Connor, O.A. Belinostat in patients with refractory or relapsed peripheral T-cell lymphoma: A perspective review. Ther. Adv. Hematol. 2015, 6, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Moore, D. Panobinostat (Farydak): A Novel Option for the Treatment of Relapsed Or Relapsed and Refractory Multiple Myeloma. Pharm. Ther. 2016, 41, 296–300. [Google Scholar]
- Lu, X.; Ning, Z.; Li, Z.; Cao, H.; Wang, X. Development of chidamide for peripheral T-cell lymphoma, the first orphan drug approved in China. Intractable Rare Dis. Res. 2016, 5, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Coiffier, B.; Pro, B.; Prince, H.M.; Foss, F.; Sokol, L.; Greenwood, M.; Caballero, D.; Borchmann, P.; Morschhauser, F.; Wilhelm, M.; et al. Results from a pivotal, open-label, phase II study of romidepsin in relapsed or refractory peripheral T-cell lymphoma after prior systemic therapy. J. Clin. Oncol. 2012, 30, 631–636. [Google Scholar] [CrossRef]
- Mackay, H.J.; Hirte, H.; Colgan, T.; Covens, A.; MacAlpine, K.; Grenci, P.; Wang, L.; Mason, J.; Pham, P.A.; Tsao, M.S.; et al. Phase II trial of the histone deacetylase inhibitor belinostat in women with platinum resistant epithelial ovarian cancer and micropapillary (LMP) ovarian tumours. Eur. J. Cancer 2010, 46, 1573–1579. [Google Scholar] [CrossRef]
- Morita, S.; Oizumi, S.; Minami, H.; Kitagawa, K.; Komatsu, Y.; Fujiwara, Y.; Inada, M.; Yuki, S.; Kiyota, N.; Mitsuma, A.; et al. Phase I dose-escalating study of panobinostat (LBH589) administered intravenously to Japanese patients with advanced solid tumors. Investig. New Drugs 2012, 30, 1950–1957. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.S.; Galloway, S.; Lagrutta, A.; Armstrong, M.; Miller, T.; Richon, V.M.; Andrews, P.A. Nonclinical safety assessment of the histone deacetylase inhibitor vorinostat. Int. J. Toxicol. 2010, 29, 3–19. [Google Scholar] [CrossRef]
- Shen, S.; Kozikowski, A.P. Why Hydroxamates May Not Be the Best Histone Deacetylase Inhibitors--What Some May Have Forgotten or Would Rather Forget? ChemMedChem 2016, 11, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Zwinderman, M.R.H.; Dekker, F.J. The Process and Strategy for Developing Selective Histone Deacetylase 3 Inhibitors. Molecules 2018, 23, 551. [Google Scholar]
- Dallavalle, S.; Pisano, C.; Zunino, F. Development and therapeutic impact of HDAC6-selective inhibitors. Biochem. Pharm. 2012, 84, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Kaletsch, A.; Pinkerneil, M.; Hoffmann, M.J.; Jaguva Vasudevan, A.A.; Wang, C.; Hansen, F.K.; Wiek, C.; Hanenberg, H.; Gertzen, C.; Gohlke, H.; et al. Effects of novel HDAC inhibitors on urothelial carcinoma cells. Clin. Epigenet. 2018, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Manzotti, G.; Ciarrocchi, A.; Sancisi, V. Inhibition of BET Proteins and Histone Deacetylase (HDACs): Crossing Roads in Cancer Therapy. Cancers 2019, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Bae, S.C. Histone deacetylase inhibitors: Molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Minucci, S.; Pelicci, P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 2006, 6, 38–51. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.P.; Singh, M.M.; Rivera-Del Valle, N.; Manton, C.A.; Chandra, J. Therapeutic strategies to enhance the anticancer efficacy of histone deacetylase inhibitors. J. Biomed. Biotechnol. 2011, 2011, 514261. [Google Scholar] [CrossRef]
- Roy, S.; Nicholson, D.W. Cross-talk in cell death signaling. J. Exp. Med. 2000, 192, 21–26. [Google Scholar] [CrossRef]
- Fulda, S. Tumor resistance to apoptosis. Int. J. Cancer 2009, 124, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.; Tu, H.C.; Kim, H.; Wang, G.X.; Bean, G.R.; Takeuchi, O.; Jeffers, J.R.; Zambetti, G.P.; Hsieh, J.J.; Cheng, E.H. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science 2010, 330, 1390–1393. [Google Scholar] [CrossRef]
- Elkholi, R.; Floros, K.V.; Chipuk, J.E. The Role of BH3-Only Proteins in Tumor Cell Development, Signaling, and Treatment. Genes Cancer 2011, 2, 523–537. [Google Scholar] [CrossRef] [Green Version]
- Matthews, G.M.; Newbold, A.; Johnstone, R.W. Intrinsic and extrinsic apoptotic pathway signaling as determinants of histone deacetylase inhibitor antitumor activity. Adv. Cancer Res. 2012, 116, 165–197. [Google Scholar] [PubMed]
- Sasca, D.; Hahnel, P.S.; Szybinski, J.; Khawaja, K.; Kriege, O.; Pante, S.V.; Bullinger, L.; Strand, S.; Strand, D.; Theobald, M.; et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood 2014, 124, 121–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hacker, S.; Karl, S.; Mader, I.; Cristofanon, S.; Schweitzer, T.; Krauss, J.; Rutkowski, S.; Debatin, K.M.; Fulda, S. Histone deacetylase inhibitors prime medulloblastoma cells for chemotherapy-induced apoptosis by enhancing p53-dependent Bax activation. Oncogene 2011, 30, 2275–2281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishioka, C.; Ikezoe, T.; Yang, J.; Udaka, K.; Yokoyama, A. Simultaneous inhibition of DNA methyltransferase and histone deacetylase induces p53-independent apoptosis via down-regulation of Mcl-1 in acute myelogenous leukemia cells. Leuk. Res. 2011, 35, 932–939. [Google Scholar] [CrossRef]
- Hirose, T.; Sowa, Y.; Takahashi, S.; Saito, S.; Yasuda, C.; Shindo, N.; Furuichi, K.; Sakai, T. p53-independent induction of Gadd45 by histone deacetylase inhibitor: Coordinate regulation by transcription factors Oct-1 and NF-Y. Oncogene 2003, 22, 7762–7773. [Google Scholar] [CrossRef] [PubMed]
- Vrana, J.A.; Decker, R.H.; Johnson, C.R.; Wang, Z.; Jarvis, W.D.; Richon, V.M.; Ehinger, M.; Fisher, P.B.; Grant, S. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene 1999, 18, 7016–7025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boucherat, O.; Chabot, S.; Paulin, R.; Trinh, I.; Bourgeois, A.; Potus, F.; Lampron, M.C.; Lambert, C.; Breuils-Bonnet, S.; Nadeau, V.; et al. HDAC6: A Novel Histone Deacetylase Implicated in Pulmonary Arterial Hypertension. Sci. Rep. 2017, 7, 4546. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Zhang, F.; Zhang, X.T.; Zhang, T.; Li, Y.H.; Fan, L.; Sun, Y.; Zhang, H.L.; Mei, Q.B. Ku70 is essential for histone deacetylase inhibitor trichostatin A-induced apoptosis. Mol. Med. Rep. 2015, 12, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.; Chen, D.J. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair 2006, 5, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, F.; Wang, T.; Flagg, T.; Deng, X. A nonhomologous end-joining pathway is required for protein phosphatase 2A promotion of DNA double-strand break repair. Neoplasia 2009, 11, 1012–1021. [Google Scholar] [CrossRef]
- Li, Z.; Owonikoko, T.K.; Sun, S.Y.; Ramalingam, S.S.; Doetsch, P.W.; Xiao, Z.Q.; Khuri, F.R.; Curran, W.J.; Deng, X. c-Myc suppression of DNA double-strand break repair. Neoplasia 2012, 14, 1190–1202. [Google Scholar] [CrossRef]
- Hada, M.; Subramanian, C.; Andrews, P.C.; Kwok, R.P. Cytosolic Ku70 regulates Bax-mediated cell death. Tumour Biol. 2016, 37, 13903–13914. [Google Scholar] [CrossRef] [Green Version]
- Antignani, A.; Youle, R.J. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr. Opin. Cell Biol. 2006, 18, 685–689. [Google Scholar] [CrossRef]
- Subramanian, C.; Opipari, A.W., Jr.; Bian, X.; Castle, V.P.; Kwok, R.P. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 2005, 102, 4842–4847. [Google Scholar] [CrossRef] [Green Version]
- Ngo, J.; Matsuyama, M.; Kim, C.; Poventud-Fuentes, I.; Bates, A.; Siedlak, S.L.; Lee, H.G.; Doughman, Y.Q.; Watanabe, M.; Liner, A.; et al. Bax deficiency extends the survival of Ku70 knockout mice that develop lung and heart diseases. Cell Death Dis. 2015, 6, e1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuyama, S.; Palmer, J.; Bates, A.; Poventud-Fuentes, I.; Wong, K.; Ngo, J.; Matsuyama, M. Bax-induced apoptosis shortens the life span of DNA repair defect Ku70-knockout mice by inducing emphysema. Exp. Biol. Med. 2016, 241, 1265–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vishnudas, V.K.; Miller, J.B. Ku70 regulates Bax-mediated pathogenesis in laminin-alpha2-deficient human muscle cells and mouse models of congenital muscular dystrophy. Hum. Mol. Genet. 2009, 18, 4467–4477. [Google Scholar] [CrossRef]
- Amsel, A.D.; Rathaus, M.; Kronman, N.; Cohen, H.Y. Regulation of the proapoptotic factor Bax by Ku70-dependent deubiquitylation. Proc. Natl. Acad. Sci. USA 2008, 105, 5117–5122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathaus, M.; Lerrer, B.; Cohen, H.Y. DeubiKuitylation: A novel DUB enzymatic activity for the DNA repair protein, Ku70. Cell Cycle 2009, 8, 1843–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perciavalle, R.M.; Opferman, J.T. Delving deeper: MCL-1’s contributions to normal and cancer biology. Trends Cell Biol. 2013, 23, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xie, M.; Li, R.; Owonikoko, T.K.; Ramalingam, S.S.; Khuri, F.R.; Curran, W.J.; Wang, Y.; Deng, X. Role of Ku70 in deubiquitination of Mcl-1 and suppression of apoptosis. Cell Death Differ. 2014, 21, 1160–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerr, E.; Holohan, C.; McLaughlin, K.M.; Majkut, J.; Dolan, S.; Redmond, K.; Riley, J.; McLaughlin, K.; Stasik, I.; Crudden, M.; et al. Identification of an acetylation-dependant Ku70/FLIP complex that regulates FLIP expression and HDAC inhibitor-induced apoptosis. Cell Death Differ. 2012, 19, 1317–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLornan, D.; Hay, J.; McLaughlin, K.; Holohan, C.; Burnett, A.K.; Hills, R.K.; Johnston, P.G.; Mills, K.I.; McMullin, M.F.; Longley, D.B.; et al. Prognostic and therapeutic relevance of c-FLIP in acute myeloid leukaemia. Br. J. Haematol. 2013, 160, 188–198. [Google Scholar] [CrossRef]
- McCann, C.; Crawford, N.; Majkut, J.; Holohan, C.; Armstrong, C.W.D.; Maxwell, P.J.; Ong, C.W.; LaBonte, M.J.; McDade, S.S.; Waugh, D.J.; et al. Cytoplasmic FLIP(S) and nuclear FLIP(L) mediate resistance of castrate-resistant prostate cancer to apoptosis induced by IAP antagonists. Cell Death Dis. 2018, 9, 1081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, H.; Yang, M.; Ye, S.; Li, L.; Zhang, H.; Hu, J.; Wang, X.; Xu, J.; Liang, A. SIRT1 inhibition impairs non-homologous end joining DNA damage repair by increasing Ku70 acetylation in chronic myeloid leukemia cells. Oncotarget 2016, 7, 13538–13550. [Google Scholar] [CrossRef] [PubMed]
- Valdez, B.C.; Li, Y.; Murray, D.; Liu, Y.; Nieto, Y.; Champlin, R.E.; Andersson, B.S. Combination of a hypomethylating agent and inhibitors of PARP and HDAC traps PARP1 and DNMT1 to chromatin, acetylates DNA repair proteins, down-regulates NuRD and induces apoptosis in human leukemia and lymphoma cells. Oncotarget 2018, 9, 3908–3921. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Nagaria, P.K.; Pawar, N.; Adewuyi, A.; Gojo, I.; Meyers, D.J.; Cole, P.A.; Rassool, F.V. Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin. Leuk. Res. 2016, 45, 14–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makita, N.; Ninomiya, I.; Tsukada, T.; Okamoto, K.; Harada, S.; Nakanuma, S.; Sakai, S.; Makino, I.; Kinoshita, J.; Hayashi, H.; et al. Inhibitory effects of valproic acid in DNA double-strand break repair after irradiation in esophageal squamous carcinoma cells. Oncol. Rep. 2015, 34, 1185–1192. [Google Scholar] [CrossRef] [Green Version]
- Rosato, R.R.; Almenara, J.A.; Maggio, S.C.; Coe, S.; Atadja, P.; Dent, P.; Grant, S. Role of histone deacetylase inhibitor-induced reactive oxygen species and DNA damage in LAQ-824/fludarabine antileukemic interactions. Mol. Cancer Ther. 2008, 7, 3285–3297. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Wang, H.; Xu, S.; Han, P.; Xin, M.; Zhou, J.L. Sensitization of suberoylanilide hydroxamic acid (SAHA) on chemoradiation for human cervical cancer cells and its mechanism. Eur. J. Gynaecol. Oncol. 2015, 36, 117–122. [Google Scholar] [PubMed]
- Busser, B.; Sancey, L.; Josserand, V.; Niang, C.; Khochbin, S.; Favrot, M.C.; Coll, J.L.; Hurbin, A. Amphiregulin promotes resistance to gefitinib in nonsmall cell lung cancer cells by regulating Ku70 acetylation. Mol. Ther. 2010, 18, 536–543. [Google Scholar] [CrossRef]
- Di Giorgio, E.; Gagliostro, E.; Brancolini, C. Selective class IIa HDAC inhibitors: Myth or reality. Cell. Mol. Life Sci. 2015, 72, 73–86. [Google Scholar] [CrossRef]
- Valenzuela-Fernandez, A.; Cabrero, J.R.; Serrador, J.M.; Sanchez-Madrid, F. HDAC6: A key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008, 18, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, N.; Amin, S.A.; Trivedi, P.; Jha, T.; Ghosh, B. HDAC3 is a potential validated target for cancer: An overview on the benzamide-based selective HDAC3 inhibitors through comparative SAR/QSAR/QAAR approaches. Eur. J. Med. Chem. 2018, 157, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Jeffers, M.; Kumar, S.; Hackett, C.; Boldog, F.; Khramtsov, N.; Qian, X.; Mills, E.; Berghs, S.C.; Carey, N.; et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem. J. 2008, 409, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, J.; Wada, T.; Shimizu, R.; Izumi, T.; Akutsu, M.; Mitsunaga, K.; Noborio-Hatano, K.; Nobuyoshi, M.; Ozawa, K.; Kano, Y.; et al. Histone deacetylases are critical targets of bortezomib-induced cytotoxicity in multiple myeloma. Blood 2010, 116, 406–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshareeda, A.T.; Negm, O.H.; Albarakati, N.; Green, A.R.; Nolan, C.; Sultana, R.; Madhusudan, S.; Benhasouna, A.; Tighe, P.; Ellis, I.O.; et al. Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res. Treat. 2013, 139, 301–310. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, X.; Shi, W.; Xu, J.; Fan, H.; Zhang, S.; Ni, R. The DNA damage repair protein Ku70 regulates tumor cell and hepatic carcinogenesis by interacting with FOXO4. Pathol. Res. Pract. 2016, 212, 153–161. [Google Scholar] [CrossRef] [PubMed]


| Substrate | HDACs | Function of Acetylation | Reference |
|---|---|---|---|
| p53 | HDAC1, 2, 3, 6, SIRT1, 2, 7 | Increases transcription activity | [33,34,35,36] |
| NF-κB | HDAC3, SIRT1, 2 | Impacts NF-κB translocation and NF-κB-mediated inflammation and transcription | [39,40,59,60] |
| Hsp90 | HDAC3, 6 | Promotes chaperone protein degradation | [49,50,51] |
| STATs | HDAC3 | Inhibits JAK2/STAT pathway and NF-kB-mediated inflammatory response | [54,61] |
| Akt | HDAC3, 6, SIRT1, 2 | Decreases Akt phosphorylation and inhibits Akt-mediated pathway | [55,56,57,58] |
| Ku70 | HDAC1, 3, 6, SIRT1, 3, 6 | Enhances DNA damage and reduces the stability of Ku70-regulated proteins | [62,63,64,65,66,67] |
| α-tubulin | HDAC6, SIRT2 | Inhibits microtubule assembly and cell motility | [68,69] |
| c-Myc | HDAC3, 6 | Inhibits the transcription ability and decreases c-Myc expression | [70,71] |
| E2F1 | HDAC1 | Increases DNA-binding affinity | [72] |
| HIF-1α | HDAC2, 4, 5, 7 | Regulates its degradation | [73,74,75,76] |
| ER | HDAC1 | Increases cell proliferation | [77] |
| β-catenin | HDAC6 | Inhibits its nuclear translocation | [78] |
| GATAs | HDAC3, 4, 5 | Affects erythroid differentiation | [79,80] |
| FoxOs | SIRT1 | Decreases DNA-binding affinity and reduces transcription activity | [81] |
| HDAC Inhibitor | Chemical Class | Structure of the Compound | HDAC Class Inhibited |
|---|---|---|---|
| Vorinostat (SAHA) | Hydroxamic acid | 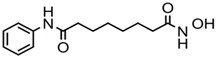 | Pan-HDACs |
| Romidepsin (FK228) | Cyclic peptide |  | Class I |
| Belinostat (PXD-101) | Hydroxamic acid |  | Pan-HDACs |
| Panobinostat (LBH-589) | Hydroxamic acid | 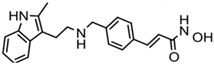 | Pan-HDACs |
| Chidamide | Benzamide |  | Class I, HDAC10 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, P.; Wang, Y.; Jing, Y. Apoptosis Induction byHistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70. Int. J. Mol. Sci. 2019, 20, 1601. https://doi.org/10.3390/ijms20071601
Gong P, Wang Y, Jing Y. Apoptosis Induction byHistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70. International Journal of Molecular Sciences. 2019; 20(7):1601. https://doi.org/10.3390/ijms20071601
Chicago/Turabian StyleGong, Ping, Yuetong Wang, and Yongkui Jing. 2019. "Apoptosis Induction byHistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70" International Journal of Molecular Sciences 20, no. 7: 1601. https://doi.org/10.3390/ijms20071601





