Identification and Characterization of EI (Elongated Internode) Gene in Tomato (Solanum lycopersicum)
Abstract
:1. Introduction
2. Results
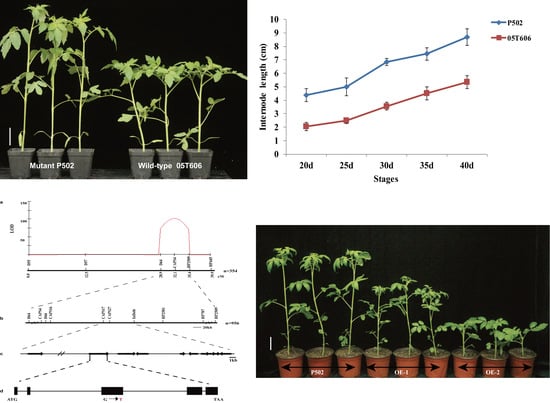
2.1. Characterization of Elongated Internode Inbred Line P502
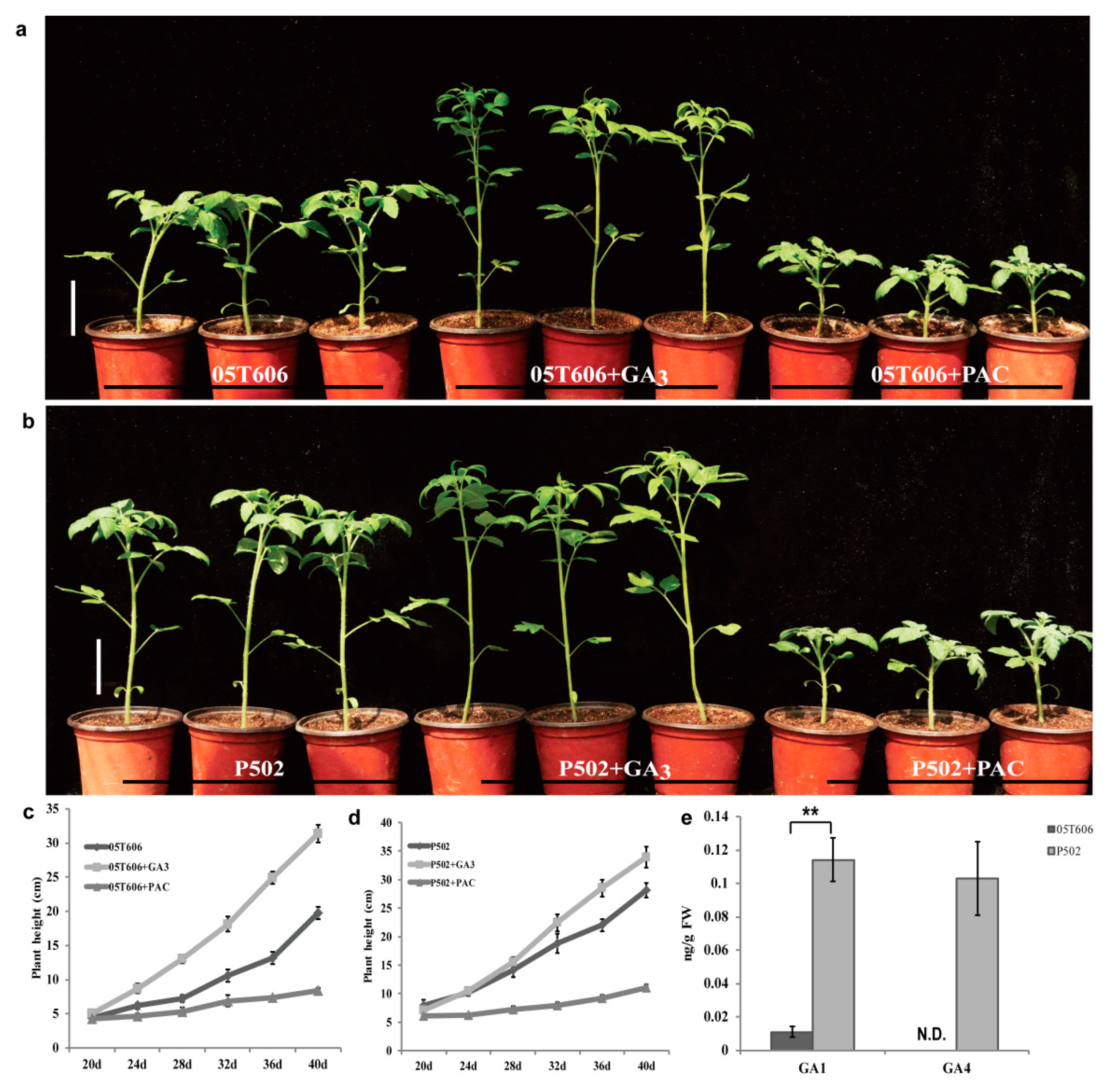
2.2. Elongated Internode Mutation Is Related to the GA Metabolic Pathway
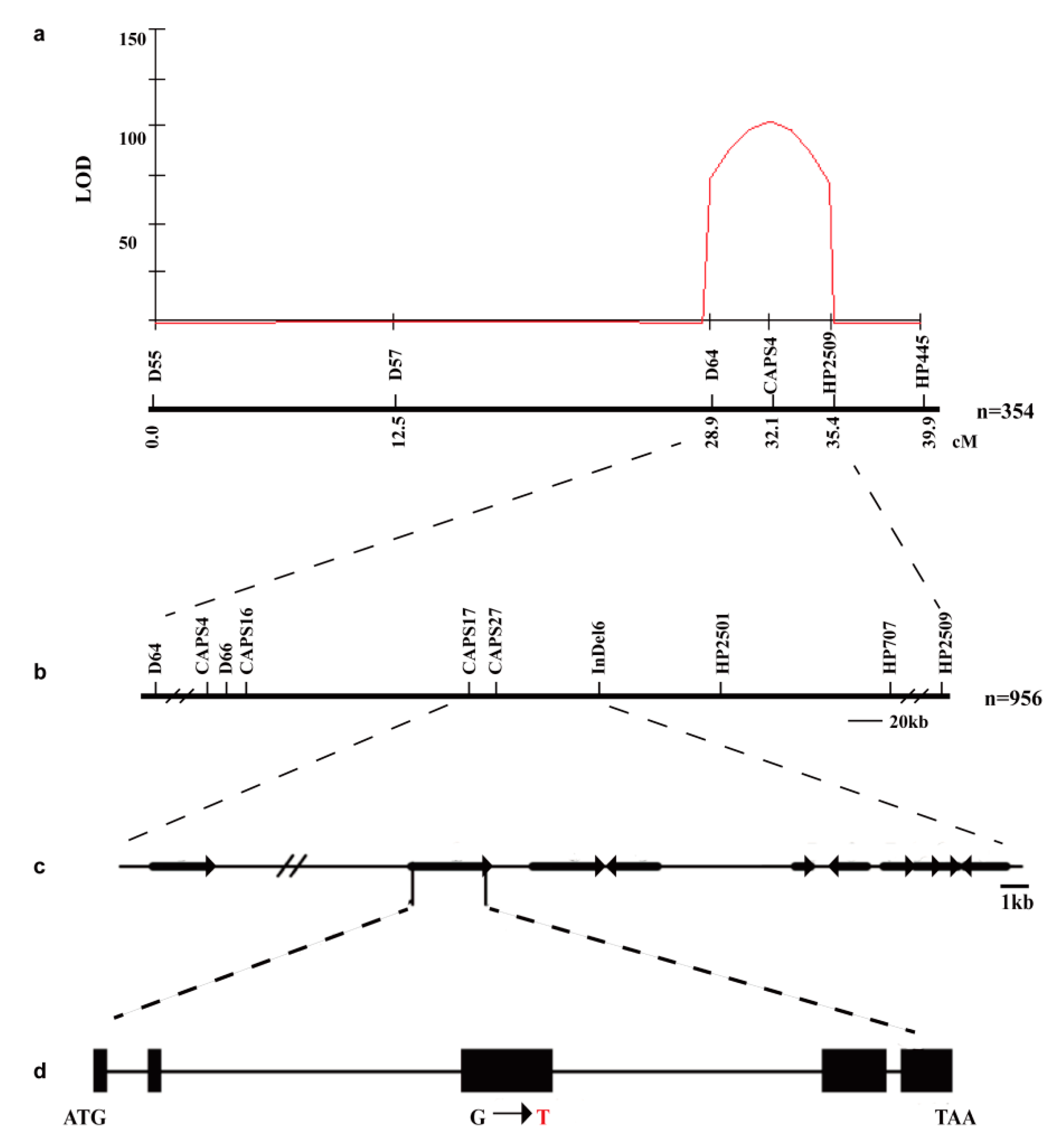
2.3. Genetic Analysis of the Elongated Internode Trait
2.4. Map-Based Cloning of the EI Gene
2.5. Protein Sequence Alignment and Phylogenetic Analysis of SlGA2ox7
2.6. Overexpression of Wild-Type EI in P502 Resulted in Dwarfism
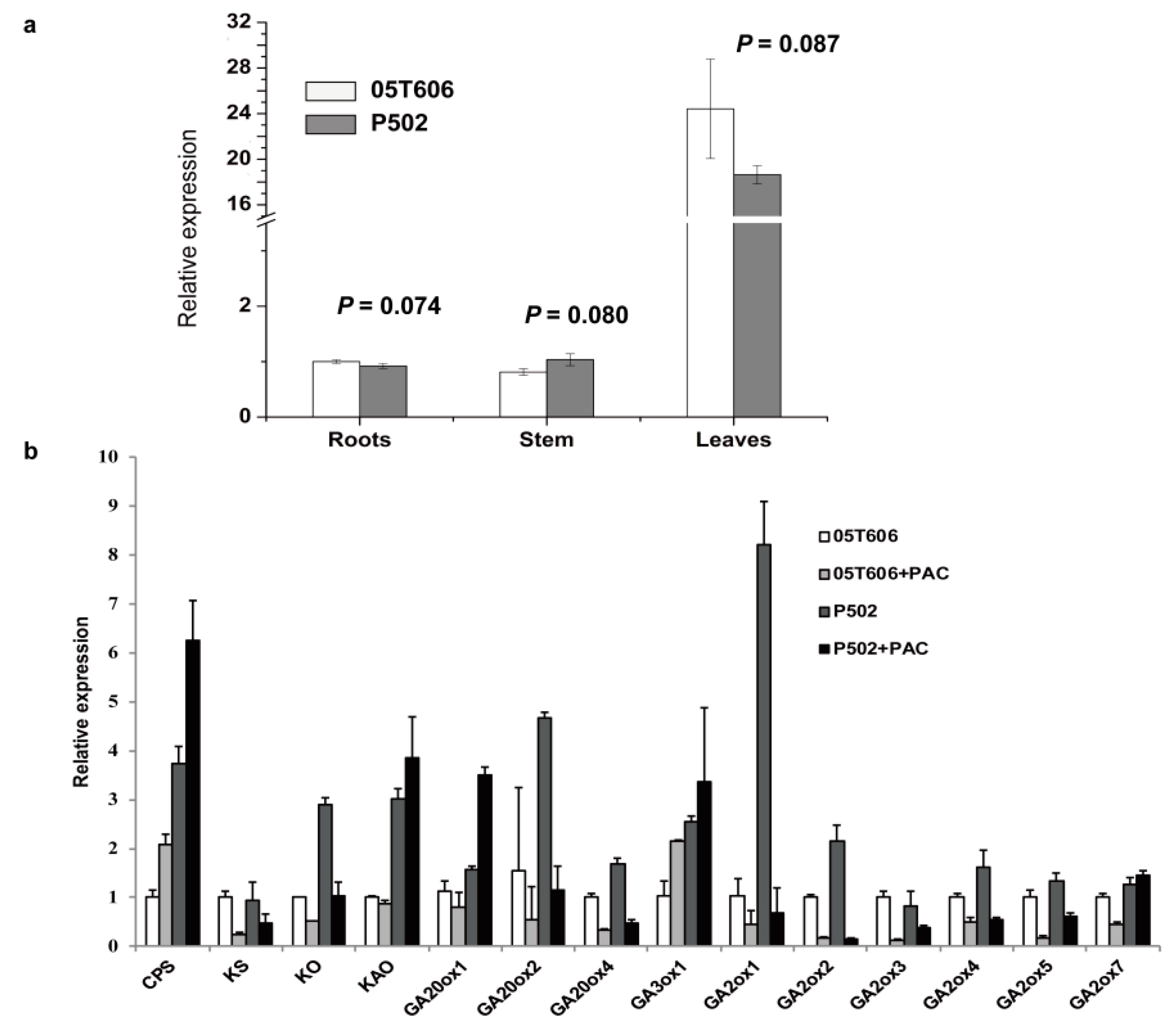
2.7. The Expression Analysis of GA Metabolic Pathway-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Scanning Electron Microscopy Observation
4.3. Exogenous GA3 Treatments and Endogenous GA Quantification
4.4. DNA Extraction and Molecular Marker Development
4.5. Mapping Strategy
4.6. RNA Extraction and qRT-PCR
4.7. Protein Sequence Alignment and Phylogenetic Analysis
4.8. Plasmid Construction and Transformation
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Salas Fernandez, G.S.; Becraft, P.W.; Yin, Y.; Lübberstedt, T. From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci. 2009, 14, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Tatsumi, T.; Sakamoto, T.; Otomo, K.; Toyomasu, T.; Kitano, H.; Ashikari, M.; Ichihara, S.; Matsuoka, M. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 2004, 54, 533–547. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Md. Babul, A.; Piao, R.; Reflinur; Md. Lutfor, R.; Yunjoo, L.; Jeonghwan, S.; Backki, K.; Hee-Jong, K. Characterization and mapping of d13, a dwarfing mutant gene, in rice. Genes Genom. 2015, 37, 893–903. [Google Scholar]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef]
- Zhou, H.; He, S.; Cao, Y.; Chen, T.; Du, B.; Chu, C.; Zhang, J.; Chen, S. OsGLU1, a putative membrane-bound endo-1,4-ß-D-glucanase from rice, affects plant internode elongation. Plant Mol. Biol. 2006, 60, 137–151. [Google Scholar] [CrossRef]
- Chen, Y.; Hou, M.; Liu, L.; Wu, S.; Shen, Y.; Ishiyama, K.; Kobayashi, M.; McCarty, D.R.; Tan, B.C. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014, 166, 2028–2039. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Z.; Ding, J.; Wu, Y.; Zhou, B.; Wang, R.; Ma, J.; Wang, S.; Zhang, X.; Xia, Z.; et al. Combined linkage and association mapping reveals QTL and candidate genes for plant and ear height in maize. Front Plant Sci. 2016, 7, 833. [Google Scholar] [CrossRef]
- Teng, F.; Zhai, L.; Liu, R.; Bai, W.; Wang, L.; Huo, D.; Tao, Y.; Zheng, Y.; Zhang, Z. ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J. 2013, 73, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wen, W.; Xie, L.; Fu, L.; Xu, D.; Fu, C.; Wang, D.; Chen, X.; Xia, X.; Chen, Q.; et al. Molecular mapping of reduced plant height gene Rht24 in Bread Wheat. Front. Plant Sci. 2017, 8, 1379. [Google Scholar] [CrossRef]
- Zhai, H.; Feng, Z.; Li, J.; Liu, X.; Xiao, S.; Ni, Z.; Sun, Q. QTL analysis of spike morphological traits and plant height in Winter Wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci. 2016, 7, 1617. [Google Scholar] [CrossRef] [PubMed]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Fujimoto, H.; Hirano, K.; Araki-Nakamura, S.; Ohmae-Shinohara, K.; Fujii, A.; Tsunashima, M.; Song, X.J.; Ito, Y.; Nagae, R.; et al. Sorghum Dw1, an agronomically important gene for lodging resistance, encodes a novel protein involved in cell proliferation. Sci. Rep. 2016, 6, 28366. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zheng, S.; Gui, J.; Fu, C.; Yu, H.; Song, D.; Shen, J.; Qin, P.; Liu, X.; Han, B.; et al. Shortened Basal Internodes encodes a gibberellin 2-oxidase and contributes to lodging resistance in rice. Mol. Plant 2018, 11, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Sheldon, C.C.; Olive, M.R.; Walker, A.R.; Zeevaart, J.A.; Peacock, W.J.; Dennis, E.S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 1998, 95, 9019–9024. [Google Scholar] [CrossRef]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 135, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, Y.; Ou, L.; Hong, H.; Wang, J.; Liu, Z.; Guo, B.; Zhang, L.; Qiu, L. Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 2018, 131, 1001–1016. [Google Scholar] [CrossRef]
- Richards, D.E.; King, K.E.; Ait-Ali, T.; Harberd, N.P. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 67–88. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Spielmeyer, W.; Ellism, M.H.; Chandler, P.M. Semidwarf (sd-1), ‘green revolution’ rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef] [PubMed]
- Koorneef, M.; Elgersma, A.; Hanhart, C.J.; Van Loenen Martinet, E.P.; Van Rijn, L.; Zeevaart, J.A. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant. 2010, 65, 33–39. [Google Scholar] [CrossRef]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Gene Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [PubMed]
- Harberd, N.P.; Freeling, M. Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics 1989, 121, 827–838. [Google Scholar]
- Winkler, R.G.; Freeling, M. Physiological genetics of the dominant gibberellin-nonresponsive maize dwarfs, Dwarf 8 and Dwarf 9. Planta 1994, 193, 341–348. [Google Scholar] [CrossRef]
- Ogawa, M.; Kusano, T.; Katsumi, M.; Sano, H. Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 2000, 245, 21–29. [Google Scholar] [CrossRef]
- Fu, X.D.; Richards, D.E.; Ait-Ali, T.; Hynes, L.W.; Ougham, H.; Peng, J.; Harberd, N.P. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef]
- Carrera, E.; Ruiz-Rivero, O.; Peres, L.E.; Atares, A.; Garcia-Martinez, J.L. Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 2012, 160, 1581–1596. [Google Scholar] [CrossRef]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1606. [Google Scholar] [CrossRef]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Foolad, M.R. Genome mapping and molecular breeding of tomato. Int. J. Plant Genom. 2007, 2007, 64358. [Google Scholar] [CrossRef] [PubMed]
- Martí, E.; Gisbert, C.; Bishop, G.J.; Dixon, M.S.; Garcíamartínez, J.L. Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 2006, 57, 2037–2047. [Google Scholar] [CrossRef] [Green Version]
- Barabaschi, D.; Tondelli, A.; Desiderio, F.; Volante, A.; Vaccino, P.; Valè, G.; Cattivelli, L. Next generation breeding. Plant Sci. 2016, 242, 3–13. [Google Scholar] [CrossRef]
- Tong, G.L.; Samuel, F.H. Fine mapping of the brachytic locus on the tomato genome. J. Amer. Soc. Hort. Sci. 2018, 143, 239–247. [Google Scholar]
- Cowling, R.J.; Kamiya, Y.; Seto, H.; Harberd, N.P. Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 1998, 117, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.A.; Solow, T.H.; Taylor, N.; Skwarecki, B.; Buels, R.; Binns, J.; Lin, C.; Wright, M.H.; Ahrens, R.; Wang, Y.; Herbst, E.V.; Keyder, E.R.; Menda, N.; Zamir, D.; Tanksley, S.D. The SOL Genomics Network: A comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005, 138, 1310–1317. [Google Scholar] [CrossRef]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef]
- Mitsunaga, S.; Tashiro, T.; Yamaguchi, J. Identification and characterization of gibberellin-insensitive mutants selected from among dwarf mutants of rice. Theor. Appl. Genet. 1994, 87, 705–712. [Google Scholar] [PubMed]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends. Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Gomi, K.; Matsuoka, M. Gibberellin signalling pathway. Curr. Opin. Plant Biol. 2003, 6, 489–493. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Zeevaart, J.A. Differential Regulation of RNA Levels of Gibberellin Dioxygenases by Photoperiod in Spinach. Plant Physiol. 2002, 130, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.N.; Proebsting, W.M.; Hedden, P. The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol. 1999, 121, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tang, D.; Shen, Y.; Qin, B.; Hong, L.; You, A.; Li, M.; Wang, X.; Yu, H.; Gu, M.; et al. Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J. Genet. Genom. 2010, 37, 23–36. [Google Scholar] [CrossRef]
- Lee, D.J.; Zeevaart, J.A. Molecular Cloning of GA 2-Oxidase3 from Spinach and Its Ectopic Expression in Nicotiana sylvestris. Plant Physiol. 2005, 138, 243–254. [Google Scholar] [CrossRef]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 2008, 6, e0103. [Google Scholar] [CrossRef]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar]
- Sun, X.R.; Liu, L.; Zhi, X.N.; Bai, J.R.; Cui, Y.N.; Shu, J.S.; Li, J.M. Genetic analysis of tomato internode length via mixed major gene plus polygene inheritance model. Sci. Hortic. 2019, 246, 759–764. [Google Scholar] [CrossRef]
- Collins, T.J. ImageJ for microscopy. BioTechniques 2007, 43, 25–30. [Google Scholar] [CrossRef] [PubMed]
- García-Hurtado, N.; Carrera, E.; Ruiz-Rivero, O.; López-Gresa, M.P.; Hedden, P.; Gong, F.; García-Martínez, J.L. The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J. Exp. Bot. 2012, 63, 5803–5813. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.F.; Gu, H.R.; Ding, C.L.; Xu, N.X.; Zhang, W.J. Effects of gibberellic acid on forage quality of rice (Oryza sativa) straw. Acta Pratacult. Sin. 2016, 25, 94–102. [Google Scholar]
- Saghaimaroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Report. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap® 4.0. Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma: Wageningen, The Netherlands, 2006. [Google Scholar]
- Van Ooijen, J.W. MapQTL® 6.0, Software for the Mapping of Quantitative trait Loci in Experimental Populations; Kyazma: Wageningen, The Netherlands, 2009. [Google Scholar]
- Li, J.; Sima, W.; Ouyang, B.; Wang, T.; Ziaf, K.; Luo, Z.; Liu, L.; Li, H.; Chen, M.; Huang, Y.; et al. Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J. Exp. Bot. 2012, 63, 6407–6420. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Madden, T.; Schäffer, A. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Sharma, M.K.; Solanke, A.U.; Jani, D.; Singh, Y.; Sharma, A.K. A simple and efficient Agrobacterium-mediated procedure for transformation of tomato. J. Biosci. 2009, 34, 423. [Google Scholar] [CrossRef] [PubMed]




| Materials | 2016 (cm) a | 2017 (cm) a | 2018 (cm) a |
|---|---|---|---|
| P1 (Heinz 1706) | 1.52 ± 0.35 a | 1.48 ± 0.63 a | 1.85 ± 0.30 a |
| P2 (P502) | 4.62 ± 0.19 c | 4.66 ± 0.60 c | 5.54 ± 0.48 c |
| F1 (Heinz 1706 × P502) | 2.74 ± 0.28 b | 2.53 ± 0.47 b | 3.10 ± 0.24 b |
| Recombinants | Genotype (F2) a | Phenotype(F3) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D64 | CAPS4 | D66 | CAPS16 | CAPS17 | CAPS27 | InDel6 | HP2501 | HP707 | HP2509 | N b | E b | |
| 6–11 | b | h | h | h | h | h | h | h | h | h | - | - |
| 13–5 | b | b | b | h | h | h | h | h | h | h | 44 | 15 |
| 7–48 | b | b | b | b | h | h | h | h | h | h | 45 | 15 |
| 7–71 | b | b | b | b | h | h | h | h | h | h | 43 | 14 |
| 15–543 | b | b | b | b | b | h | h | h | h | h | 45 | 14 |
| 17–18 | b | b | b | b | b | h | h | h | h | h | 45 | 15 |
| 15–741 | h | h | h | h | h | h | b | b | b | b | 46 | 14 |
| 1–2 | h | h | h | h | h | h | b | b | b | b | 45 | 15 |
| 1–72 | h | h | h | h | h | h | h | b | b | b | 44 | 16 |
| 6–21 | h | h | h | h | h | h | h | h | h | b | - | - |
| Gene ID a | Position | Functional Annotation |
|---|---|---|
| solyc02g080110.2 | SL2.50ch02: 44414686..44417879 (+) | Unknown Protein (AHRD V1) |
| solyc02g080120.1 | SL2.50ch02: 44432042..44436667 (+) | Gibberellin 2-beta-dioxygenase 7 |
| solyc02g080130.2 | SL2.50ch02: 44439349..44443883 (+) | Chaperone dnaj-like protein |
| solyc02g080140.2 | SL2.50ch02: 44444670..44447584 (-) | cysteine-rich PDZ-binding protein |
| solyc02g080150.1 | SL2.50ch02: 44458406..44458810 (+) | uncharacterized LOC101262168 |
| solyc02g080160.2 | SL2.50ch02: 44460578..44462486 (-) | probable xyloglucan endotransglucosylase/hydrolase protein 8 |
| solyc02g080170.1 | SL2.50ch02: 44471643..44473397 (+) | pentatricopeptide repeat-containing protein At4g21170 |
| solyc02g080180.1 | SL2.50ch02: 44474353..44475381 (+) | Probable dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 3B |
| solyc02g080190.2 | SL2.50ch02: 44477570..44478612 (+) | Nuclear transport factor 2 |
| solyc02g080200.2 | SL2.50ch02: 44478656..44480593 (-) | pectinesterase-like |
| Primer Name | Forward Primer (5′-3′) | Reverse primer (5′-3′) | Type | Enzyme |
|---|---|---|---|---|
| D55 | AATGACTTACCTACTGGAAAGC | GATTGATCACCCTTTGGATA | InDel | |
| D57 | GAGACATCACTTTGCCTTTC | AAAAGTCTCTCCGCCTATGT | InDel | |
| D64 | TTGTTACCGCTTACTTTGGT | CACAGCTGTTGATTTCTTCA | InDel | |
| CAPS4 | GCATTGCAACCTATTCTCAC | TCTGTAGTTTCCGTCTTCTT | CAPS | HaeIII |
| D66 | CGTTGTCTAGGTCAATAGCC | AGGTGTTACACTTTCTACGTCT | InDel | |
| CAPS16 | AGAGAAGGAGGATTCGGGTT | ATAGGGGCATTATCAAAAGG | CAPS | BsrDI |
| CAPS17 | TAAGTTAGCCATATAAAAC | AAATGACACAGCGAGACA | CAPS | MboII |
| CAPS27 | GAGAAAATTATTTGGGATAC | ATTAAAACTTTGATGCCTAC | CAPS | MfeI |
| InDel6 | ACAATCCCAGTTTATGTGAT | ATATTTGGTGTTTTCTGTTT | InDel | |
| HP2501 | CTTTTCACAAAACTAACACAGG | TGACAATATAAGCATTTGTCGC | InDel | |
| HP707 | TCCGATGTAACATCACGCAA | GTTGATCACCTTCAGACAGC | InDel | |
| D67 | AGCTTTTATAGCACGTACCG | CCATACTCTACTTATGCTGCAA | InDel | |
| HP2509 | ACCTCGACACTGGTTCACTC | GTGACTCATATACACCCTTACCTA | InDel | |
| HP445 | GAGAACATCTGTACCAGCCT | CAAGTATCTATATGCCTGACAAC | InDel |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Shu, J.; Ali Mohamed, A.M.; Deng, X.; Zhi, X.; Bai, J.; Cui, Y.; Lu, X.; Du, Y.; Wang, X.; et al. Identification and Characterization of EI (Elongated Internode) Gene in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2019, 20, 2204. https://doi.org/10.3390/ijms20092204
Sun X, Shu J, Ali Mohamed AM, Deng X, Zhi X, Bai J, Cui Y, Lu X, Du Y, Wang X, et al. Identification and Characterization of EI (Elongated Internode) Gene in Tomato (Solanum lycopersicum). International Journal of Molecular Sciences. 2019; 20(9):2204. https://doi.org/10.3390/ijms20092204
Chicago/Turabian StyleSun, Xiaorong, Jinshuai Shu, Ali Mohamed Ali Mohamed, Xuebin Deng, Xiaona Zhi, Jinrui Bai, Yanan Cui, Xiaoxiao Lu, Yongchen Du, Xiaoxuan Wang, and et al. 2019. "Identification and Characterization of EI (Elongated Internode) Gene in Tomato (Solanum lycopersicum)" International Journal of Molecular Sciences 20, no. 9: 2204. https://doi.org/10.3390/ijms20092204