Phenolic Compounds with Antioxidant Properties from Canola Meal Extracts Inhibit Adipogenesis
Abstract
:1. Introduction
2. Results
2.1. Phenolic Composition of Canola Meal Extracts
2.2. Canola Meal Extracts Inhibit Intracellular Lipid Accumulation
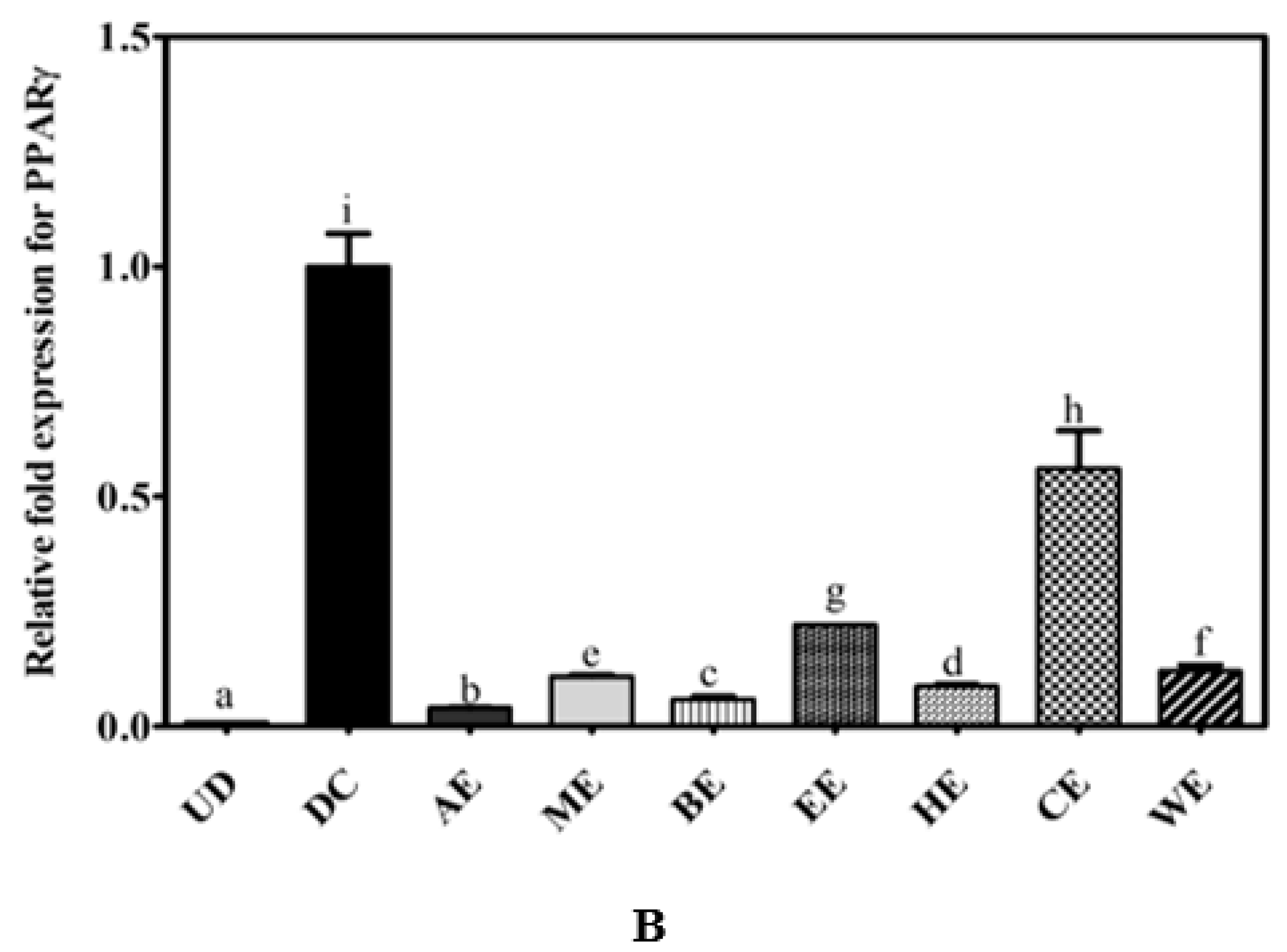
2.3. Canola Meal Extract Mediated Reductions in Lipid Accumulation are Correlated with a Reduction in PPARγ Expression
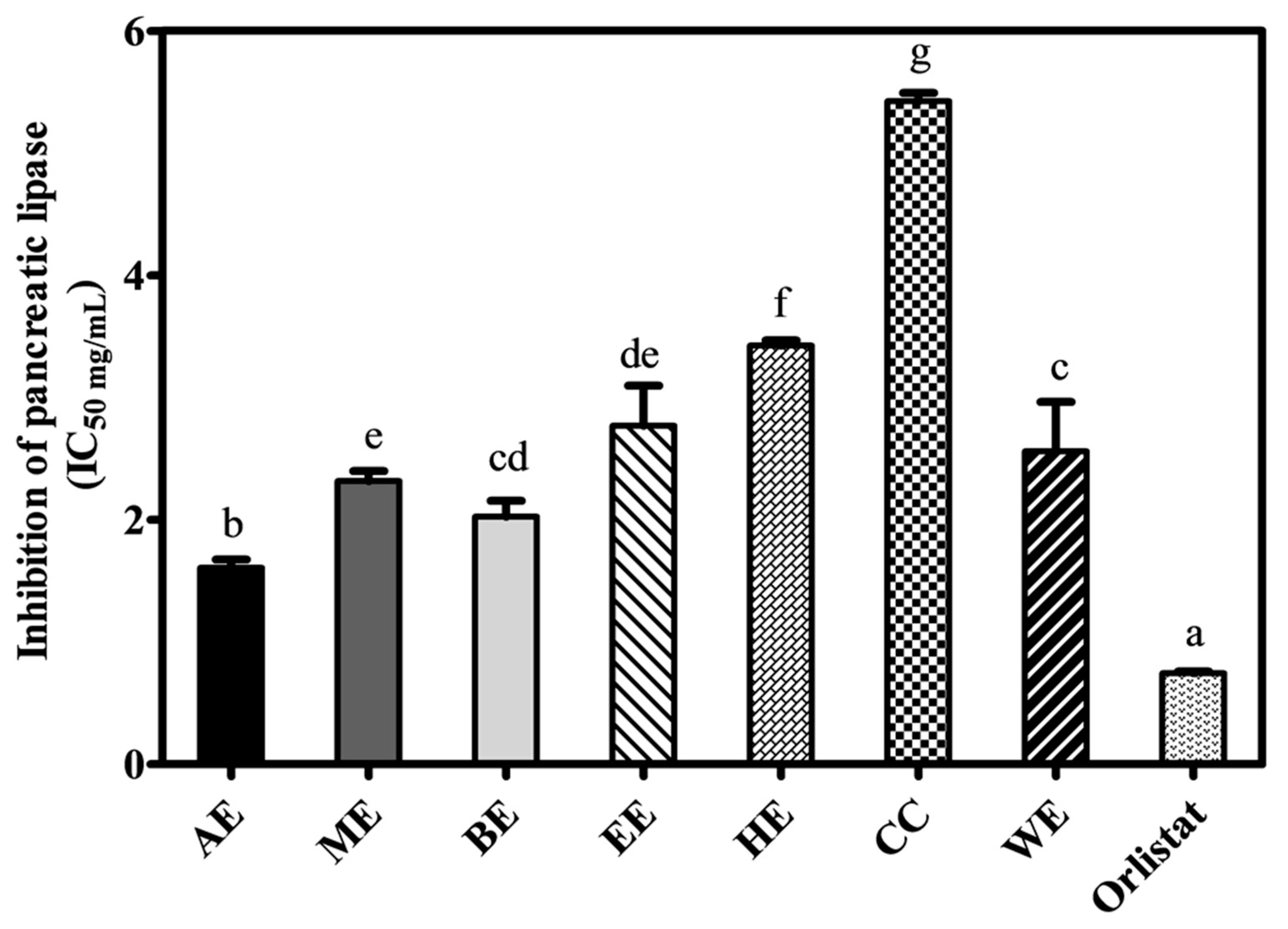
2.4. Inhibition of Pancreatic Lipase Activity
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Recovery of Canola Meal Biophenols
4.3. Measurement of Total Phenolic Content
4.4. Cell Culture and Adipogenic Differentiation
4.5. Oil-Red O Staining Staining and Quantification of Intracellular Lipid Droplets
4.6. Immunofluorescence Staining of PPARγ
4.7. Quantitative PCR (qPCR) of PPARγ Gene Expression
4.8. Pancreatic Lipase Inhibition
4.9. Chromatographic Characterization of Canola Extracts
4.9.1. Online High-Performance Liquid Chromatography with Diode Array Detector Coupled with ABTS Scavenging Activity (HPLC-DAD-Online ABTS).
4.9.2. High-Performance Liquid Chromatography-Diode Array Detection--Tandem Mass (HPLC-DAD-MS/MS).
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahman, M.; Khatun, A.; Liu, L.; Barkla, B.J. Brassicaceae mustards: Traditional and agronomic uses in Australia and New Zealand. Molecules 2018, 23, 231. [Google Scholar] [CrossRef] [Green Version]
- Obied, H.K.; Song, Y.; Foley, S.; Loughlin, M.; Rehman, A.-U.; Mailer, R.; Masud, T.; Agboola, S. Biophenols and antioxidant properties of australian canola meal. J. Agric. Food Chem. 2013, 61, 9176–9184. [Google Scholar] [CrossRef]
- Tangney, C.C.; Rasmussen, H.E. Polyphenols, inflammation, and cardiovascular disease. Curr. Atheroscler. Rep. 2013, 15, 324. [Google Scholar] [CrossRef]
- Hollman, P.C.; Cassidy, A.; Comte, B.; Heinonen, M.; Richelle, M.; Richling, E.; Serafini, M.; Scalbert, A.; Sies, H.; Vidry, S. The biological relevance of direct antioxidant effects of polyphenols for cardiovascular health in humans is not established. J. Nutr. 2011, 141, 989S–1009S. [Google Scholar] [CrossRef] [Green Version]
- Choy, K.W.; Murugan, D.D.; Leong, X.-F.; Abas, R.; Alias, A. Flavonoids as natural anti-inflammatory agents targeting nuclear factor-kappa B (NFκB) signalling in cardiovascular diseases: A mini review. Front. Pharmacol. 2019, 10, 1295. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-O.; Lee, C.Y. Comprehensive study on vitamin C equivalent antioxidant capacity (VCEAC) of various polyphenolics in scavenging a free radical and its structural relationship. Crit. Rev. Food Sci. Nutr. 2004, 44, 253–273. [Google Scholar] [CrossRef]
- Csiszar, A.; Smith, K.; Labinskyy, N.; Orosz, Z.; Rivera, A.; Ungvari, Z. Resveratrol attenuates TNF-α-induced activation of coronary arterial endothelial cells: Role of NF-κB inhibition. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1694–H1699. [Google Scholar] [CrossRef] [PubMed]
- Engler, M.B.; Engler, M.M.; Chen, C.Y.; Malloy, M.J.; Browne, A.; Chiu, E.Y.; Kwak, H.-K.; Milbury, P.; Paul, S.M.; Blumberg, J. Flavonoid-rich dark chocolate improves endothelial function and increases plasma epicatechin concentrations in healthy adults. J. Am. Coll. Nutr. 2004, 23, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.J.; Dwyer, J.T.; Jacques, P.F.; McCullough, M.L. Associations between flavonoids and cardiovascular disease incidence or mortality in European and US populations. Nutr. Rev. 2012, 70, 491–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosso, G.; Micek, A.; Godos, J.; Pajak, A.; Sciacca, S.; Galvano, F.; Giovannucci, E.L. Dietary flavonoid and lignan intake and mortality in prospective cohort studies: Systematic review and dose-response meta-analysis. Am. J. Epidemiol. 2017, 185, 1304–1316. [Google Scholar] [CrossRef]
- Kumar, S.; Andy, A. Health promoting bioactive phytochemicals from Brassica. Int. Food Res. J. 2012, 19, 141–152. [Google Scholar]
- Amorati, R.; Valgimigli, L. Methods to measure the antioxidant activity of phytochemicals and plant extracts. J. Agric. Food Chem. 2018, 66, 3324–3329. [Google Scholar] [CrossRef] [PubMed]
- Aider, M.; Barbana, C. Canola proteins: Composition, extraction, functional properties, bioactivity, applications as a food ingredient and allergenicity - A practical and critical review. Trends Food Sci. Technol. 2011, 22, 21–39. [Google Scholar] [CrossRef]
- Alashi, A.M.; Blanchard, C.L.; Mailer, R.J.; Agboola, S.O.; Mawson, A.J.; Aluko, R.E.; Strappe, P. Effects of canola proteins and hydrolysates on adipogenic differentiation of C3H10T/2 mesenchymal stem cells. Food Chem. 2015, 185, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Hocking, S.; Dear, A.; Cowley, M.A. Current and emerging pharmacotherapies for obesity in Australia. Obes. Res. Clin. Pract. 2017, 11, 501–521. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, C.; Zhao, S.; Ge, F.; Liu, D. The inhibitory effect of different solvents extracts from walnut shell (Juglans regia L.) on pancreatic lipase and adipogenesis of 3T3-L1 preadipocytes. J. Food Nutr. Res. 2014, 2, 664–670. [Google Scholar] [CrossRef] [Green Version]
- Rayalam, S.; Della-Fera, M.A.; Baile, C.A. Phytochemicals and regulation of the adipocyte life cycle. J. Nutr. Biochem. 2008, 19, 717–726. [Google Scholar] [CrossRef]
- Marseglia, L.; Manti, S.; D’Angelo, G.; Nicotera, A.; Parisi, E.; Di Rosa, G.; Gitto, E.; Arrigo, T. Oxidative stress in obesity: A critical component in human diseases. Int. J. Mol. Sci. 2015, 16, 378–400. [Google Scholar] [CrossRef] [Green Version]
- Soeng, S.; Evacuasiany, E.; Widowati, W.; Fauziah, N.; Manik, V.; Maesaroh, M. Inhibitory potential of rambutan seeds extract and fractions on adipogenesis in 3T3-L1 cell line. J. Exp. Integr. Med. 2015, 5, 55–60. [Google Scholar] [CrossRef]
- Sergent, T.; Vanderstraeten, J.; Winand, J.; Beguin, P.; Schneider, Y.-J. Phenolic compounds and plant extracts as potential natural anti-obesity substances. Food Chem. 2012, 135, 68–73. [Google Scholar] [CrossRef]
- Stephens, J.M. The fat controller: Adipocyte development. PLoS Biol. 2012, 10, e1001436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalska, K.; Olejnik, A.; Rychlik, J.; Grajek, W. Cranberries (Oxycoccus quadripetalus) inhibit adipogenesis and lipogenesis in 3T3-L1 cells. Food Chem. 2014, 148, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Callcott, E.T.; Santhakumar, A.B.; Strappe, P.; Luo, J.; Blanchard, C.L. Polyphenols from Australian-grown pigmented red and purple rice inhibit adipocyte differentiation. J. Cereal Sci. 2018, 81, 140–146. [Google Scholar] [CrossRef]
- Lee, H.; Li, H.; Kweon, M.; Choi, Y.; Kim, M.; Ryu, J.-H. Isobavachalcone from angelica keiskei inhibits adipogenesis and prevents lipid accumulation. Int. J. Mol. Sci. 2018, 19, 1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chayaratanasin, P.; Caobi, A.; Suparpprom, C.; Saenset, S.; Pasukamonset, P.; Suanpairintr, N.; Barbieri, M.A.; Adisakwattana, S. Clitoria ternatea flower petal extract inhibits adipogenesis and lipid accumulation in 3T3-L1 preadipocytes by downregulating adipogenic gene expression. Molecules 2019, 24, 1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, H.; McGhie, T.K.; Bentley-Hewitt, K.; Christeller, J. PPARγ as a sensor of lipase activity and a target for the lipase inhibitor orlistat. Lipids Health Dis. 2013, 12, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, H.; Imran, K.; Rahman, S.; Yoon, D.; Marimuthu, V.; Kim, Y. Sinapic Acid Induces the Expression of Thermogenic Signature Genes and Lipolysis through Activation of Pka/Creb Signaling in Brown Adipocytes. BMB Report. Available online: https://europepmc.org/article/med/31401979 (accessed on 12 August 2019).
- Alam, M.A.; Subhan, N.; Hossain, H.; Hossain, M.; Reza, H.M.; Rahman, M.M.; Ullah, M.O. Hydroxycinnamic acid derivatives: A potential class of natural compounds for the management of lipid metabolism and obesity. Nutr. Metab. 2016, 13, 27. [Google Scholar] [CrossRef] [Green Version]
- Aranaz, P.; Navarro-Herrera, D.; Zabala, M.; Miguéliz, I.; Romo-Hualde, A.; López-Yoldi, M.; Martínez, J.A.; Vizmanos, J.L.; Milagro, F.I.; González-Navarro, C.J. Phenolic compounds inhibit 3T3-L1 adipogenesis depending on the stage of differentiation and their binding affinity to PPARγ. Molecules 2019, 24, 1045. [Google Scholar] [CrossRef] [Green Version]
- Goh, S.; Kim, D.; Choi, M.-H.; Shin, H.-J.; Kwon, S. Effects of bamboo stem extracts on adipogenic differentiation and lipid metabolism regulating genes. Biotechnol. Bioprocess Eng. 2019, 24, 454–463. [Google Scholar] [CrossRef]
- Sferrazzo, G.; Palmeri, R.; Vanella, L.; Parafati, L.; Ronsisvalle, S.; Biondi, A.; Basile, F.; Li Volti, G.; Barbagallo, I. Mangifera indica L. leaf extract induces adiponectin and regulates adipogenesis. Int. J. Mol. Sci. 2019, 20, 3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaiittianan, R.; Sutthanut, K.; Rattanathongkom, A. Purple corn silk: A potential anti-obesity agent with inhibition on adipogenesis and induction on lipolysis and apoptosis in adipocytes. J. Ethnopharmacol. 2017, 201, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; de Mejia, E.G.; Luna-Vital, D.; Tao, T.; Chandrasekaran, S.; Chatham, L.; Juvik, J.; Singh, V.; Kumar, D. Relationship of phenolic composition of selected purple maize (Zea mays L.) genotypes with their anti-inflammatory, anti-adipogenic and anti-diabetic potential. Food Chem. 2019, 289, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Les, F.; Arbonés-Mainar, J.M.; Valero, M.S.; López, V. Pomegranate polyphenols and urolithin A inhibit α-glucosidase, dipeptidyl peptidase-4, lipase, triglyceride accumulation and adipogenesis related genes in 3T3-L1 adipocyte-like cells. J. Ethnopharmacol. 2018, 220, 67–74. [Google Scholar] [CrossRef]
- Hussain, S. Bioactive Compounds in Canola Meal. Ph.D. Thesis, Charles Sturt University, Bathurst, Australia, 2015. [Google Scholar]
- Gosmann, G.; Barlette, A.G.; Dhamer, T.; Arçari, D.P.; Santos, J.C.; de Camargo, E.R.; Acedo, S.; Gambero, A.; Gnoatto, S.C.B.; Ribeiro, M.L. Phenolic compounds from mate (Ilex paraguariensis) inhibit adipogenesis in 3T3-L1 preadipocytes. Plant Foods Hum. Nutr. 2012, 67, 156–161. [Google Scholar] [CrossRef]
- Greenberg, A.S.; Coleman, R.A.; Kraemer, F.B.; McManaman, J.L.; Obin, M.S.; Puri, V.; Yan, Q.-W.; Miyoshi, H.; Mashek, D.G. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Investig. 2011, 121, 2102–2110. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.P.; Kaneda, T.; Morita, H. Plant natural products as an anti-lipid droplets accumulation agent. J. Nat. Med. 2014, 68, 253–266. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Do, H.-J.; Kim, O.Y.; Shin, M.-J. Antiobesity effects of quercetin-rich onion peel extract on the differentiation of 3T3-L1 preadipocytes and the adipogenesis in high fat-fed rats. Food Chem. Toxicol. 2013, 58, 347–354. [Google Scholar] [CrossRef]
- Seyedan, A.; Alshawsh, M.A.; Alshagga, M.A.; Koosha, S.; Mohamed, Z. Medicinal plants and their inhibitory activities against pancreatic lipase: A review. Evid. Based Complement. Altern. Med. 2015, 2015, 973143. [Google Scholar] [CrossRef] [Green Version]
- Azmir, J.; Zaidul, I.S.M.; Rahman, M.M.; Sharif, K.M.; Mohamed, A.; Sahena, F.; Jahurul, M.H.A.; Ghafoor, K.; Norulaini, N.A.N.; Omar, A.K.M. Techniques for extraction of bioactive compounds from plant materials: A review. J. Food Eng. 2013, 117, 426–436. [Google Scholar] [CrossRef]
- Warnke, I.; Goralczyk, R.; Fuhrer, E.; Schwager, J. Dietary constituents reduce lipid accumulation in murine C3H10 T1/2 adipocytes: A novel fluorescent method to quantify fat droplets. Nutr. Metab. 2011, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karmase, A.; Jagtap, S.; Bhutani, K.K. Anti adipogenic activity of Aegle marmelos Correa. Phytomedicine 2013, 20, 1267–1271. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Diosady, L. Interactions between canola proteins and phenolic compounds in aqueous media. Food Res. Int. 2000, 33, 725–731. [Google Scholar] [CrossRef]
- Barthet, V.J. Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Przybylski, R.; Eskin, N.M. Oil composition and properties. In Canola; Elsevier: Amsterdam, The Netherlands, 2011; pp. 189–227. [Google Scholar]
- Obied, H.K. Biography of biophenols: Past, present and future. Funct. Foods Health Dis. 2013, 3, 230–241. [Google Scholar] [CrossRef]
- Soto, C.; Caballero, E.; Pérez, E.; Zúñiga, M.E. Effect of extraction conditions on total phenolic content and antioxidant capacity of pretreated wild Peumus boldus leaves from Chile. Food Bioprod. Process. 2014, 92, 328–333. [Google Scholar] [CrossRef]
- Babbar, N.; Oberoi, H.S.; Sandhu, S.K.; Bhargav, V.K. Influence of different solvents in extraction of phenolic compounds from vegetable residues and their evaluation as natural sources of antioxidants. J. Food Sci. Technol. 2014, 51, 2568–2575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ignat, I.; Volf, I.; Popa, V.I. A critical review of methods for characterisation of polyphenolic compounds in fruits and vegetables. Food Chem. 2011, 126, 1821–1835. [Google Scholar] [CrossRef]
- Singh, R.; Artaza, J.N.; Taylor, W.E.; Gonzalez-Cadavid, N.F.; Bhasin, S. Androgens stimulate myogenic differentiation and inhibit adipogenesis in C3H 10T1/2 pluripotent cells through an androgen receptor-mediated pathway. Endocrinology 2003, 144, 5081–5088. [Google Scholar] [CrossRef]
- Thyagarajan-Sahu, A.; Lane, B.; Sliva, D. ReishiMax, mushroom based dietary supplement, inhibits adipocyte differentiation, stimulates glucose uptake and activates AMPK. BMC Complement. Altern. Med. 2011, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Nakai, M.; Fukui, Y.; Asami, S.; Toyoda-Ono, Y.; Iwashita, T.; Shibata, H.; Mitsunaga, T.; Hashimoto, F.; Kiso, Y. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J. Agric. Food Chem. 2005, 53, 4593–4598. [Google Scholar] [CrossRef]






| Peak No. | Identity | AE | RT | λmax | ABTS | ESI+ | ESI- | MW |
|---|---|---|---|---|---|---|---|---|
| 1 | Sinapine | ✓✓✓ | 15.7 | 237, 328 | ++ | 310 | 294, 663 | 310 |
| 2 | Ferroyl choline(4-0-8’) guiacyl | ✓✓ | 17.7 | 270b, 325b | +++ | 476 | NI | 476 |
| 3 | Unknown | ✘ | 24.6 | 260s, 280s, 290s | ++ | 429 | 427 | 428 |
| 4 | Ferroyl choline guiacyl isomer | ✓ | 18.3 | 270b, 325b | ++ | 476 | NI | 476 |
| 5 | Kaempherol-sinapoyl-trihexoside | ✓✓ | 26.3 | 268, 333 | + | 979 | 978 | 977 |
| 6 | Trans-sinapic acid | ✓✓ | 32.6 | 324 | ++ | NI | 223 | 224 |
| 7 | Feruloyl choline (4-O-8’) guaiacyl-di-sinapoyl | ✓✓ | 36.1 | 323 | ++ | 682 | NI | 682 |
| 8 | Disinapoyl dihexoside | ✓✓ | 38.6 | 230, 330 | ++ | NI | 753 | 754 |
| 9 | Disinapoyl hexoside | ✓✓ | 40.7 | 330 | ++ | NI | 591 | 592 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.; Rehman, A.U.; Luckett, D.J.; Blanchard, C.L.; Obied, H.K.; Strappe, P. Phenolic Compounds with Antioxidant Properties from Canola Meal Extracts Inhibit Adipogenesis. Int. J. Mol. Sci. 2020, 21, 1. https://doi.org/10.3390/ijms21010001
Hussain S, Rehman AU, Luckett DJ, Blanchard CL, Obied HK, Strappe P. Phenolic Compounds with Antioxidant Properties from Canola Meal Extracts Inhibit Adipogenesis. International Journal of Molecular Sciences. 2020; 21(1):1. https://doi.org/10.3390/ijms21010001
Chicago/Turabian StyleHussain, Saira, Ata Ur Rehman, David J. Luckett, Christopher L. Blanchard, Hassan K. Obied, and Padraig Strappe. 2020. "Phenolic Compounds with Antioxidant Properties from Canola Meal Extracts Inhibit Adipogenesis" International Journal of Molecular Sciences 21, no. 1: 1. https://doi.org/10.3390/ijms21010001






