Galactofuranose-Related Enzymes: Challenges and Hopes
Abstract
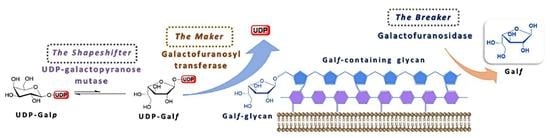
:1. Introduction
2. Occurrence in Nature
2.1. In Bacteria
2.2. In Fungi
2.3. In Protozoa
2.4. In Other Organisms
3. Aspects of Enzymatic Biosynthesis and Metabolism
3.1. UDP-Galactopyranose Mutase (UGM)
3.2. Galactofuranosyltransferase (GalfT)
3.3. Galactofuranosidases
4. Galactofuranose Antigens—Therapeutic and Diagnostic Target
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ELISA | enzyme-linked immunosorbent assay |
| EPS | exopolysaccharides |
| EPS | extracellular polysaccharides |
| FAD | flavine adenine dinucleotide |
| Galf | galactofuranose |
| GalfT | galactofuranosyltransferase |
| Galp | galactopyranose |
| GIPLs | glycoinosinositolphospholipids |
| GM | galactomannan |
| HS toxin | host-selective toxin |
| LPGs | lipophosphoglycans |
| LPPG | lipopeptidophosphoglycan |
| LPS | lipopolysaccharide |
| ORF | open reading frame |
| PAMPs | pathogen-associated molecular patterns |
| PDB | Protein Data Bank |
| PET/MR | positron emission tomography/magnetic resonance |
| pNP-β-d-Galf | para-nitrophenyl β-d-galactofuranose |
| pPGM | peptidophosphogalactomannan |
| UDP | uracil diphosphate |
| UDP-Galf | uracil diphosphate galactofuranose |
| UGM | UDP-galactopyranose mutase |
References
- Marino, C.; de Lederkremer, R.M. Galactose Configurations in Nature with Emphasis on the Biosynthesis of Galactofuranose in Glycans. In Galactose: Structure and Function in Biology and Medicine, 1st ed.; Pomin, V.H., Ed.; Nova Science Publisher Inc.: Hauppauge, NY, USA, 2014; Volume 2, pp. 107–133. [Google Scholar]
- van Rantwijk, F. Monosaccharides. Their Chemistry and Their Roles in Natural Products. Recl. Trav. Chim. Pays-Bas 1996, 115, 420. [Google Scholar] [CrossRef]
- Tefsen, B.; Ram, A.F.; van Die, I.; Routier, F.H. Galactofuranose in Eukaryotes: Aspects of Biosynthesis and Functional Impact. Glycobiology 2012, 22, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Peltier, P.; Euzen, R.; Daniellou, R.; Nugier-Chauvin, C.; Ferrières, V. Recent Knowledge and Innovations Related to Hexofuranosides: Structure, Synthesis and Applications. Carbohydr. Res. 2008, 343, 1897–1923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, C.; Gallo-Rodriguez, C.; de Lederkremer, R.M. Galactofuranosyl-Containing Glycans: Occurrence, Synthesis and Biochemistry. In Glycans: Biochemistry, Characterization and Applications, 1st ed.; Mora-Montes, H.M., Ed.; Nova Science Publisher Inc.: Hauppauge, NY, USA, 2012; pp. 207–268. [Google Scholar]
- Eppe, G.; Bkassiny, S.E.; Vincent, S.P. Galactofuranose Biosynthesis: Discovery, Mechanisms and Therapeutic Relevance. In Carbohydrates in Drug Design and Discovery; Jiménez-Barbero, J., Cañada, F.J., Martín-Santamaría, S., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2015; pp. 209–241. [Google Scholar] [CrossRef]
- Haworth, W.N.; Porter, C.R. Isolation of Crystalline α- and β-Ethylglucofuranosides (γ-Ethylglucosides) and Other Crystalline Derivatives of Glucofuranose. J. Chem. Soc. Resumed 1929, 2796–2806. [Google Scholar] [CrossRef]
- Clutterbuck, P.W.; Haworth, W.N.; Raistrick, H.; Smith, G.; Stacey, M. Studies in the Biochemistry of Micro-Organisms. Biochem. J. 1934, 28, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Haworth, W.N.; Raistrick, H.; Stacey, M. Polysaccharides Synthesised by Micro-Organisms. Biochem. J. 1937, 31, 640–644. [Google Scholar] [CrossRef] [Green Version]
- Green, J.W.; Pacsu, E. Glycofuranosides and Thioglycofuranosides. III. New Crystalline Furanosides of d-Galactose and l-Arabinose. J. Am. Chem. Soc. 1938, 60, 2056–2057. [Google Scholar] [CrossRef]
- Lowary, T.L. Twenty Years of Mycobacterial Glycans: Furanosides and Beyond. Acc. Chem. Res. 2016, 49, 1379–1388. [Google Scholar] [CrossRef]
- Thanna, S.; Sucheck, S.J. Targeting the Trehalose Utilization Pathways of Mycobacterium Tuberculosis. MedChemComm 2016, 7, 69–85. [Google Scholar] [CrossRef] [Green Version]
- Jankute, M.; Cox, J.A.G.; Harrison, J.; Besra, G.S. Assembly of the Mycobacterial Cell Wall. Annu. Rev. Microbiol. 2015, 69, 405–423. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the Wall: Extracellular Vesicles in Gram-Positive Bacteria, Mycobacteria and Fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrahams, K.A.; Besra, G.S. Mycobacterial Cell Wall Biosynthesis: A Multifaceted Antibiotic Target. Parasitology 2018, 145, 116–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaoka, M.; Hashimoto, S.; Shibata, H.; Kimura, I.; Kimura, K.; Sawada, H.; Yokokura, T. Structure of a Galactan from Cell Walls of Bifidobacterium Catenulatum YIT4016. Carbohydr. Res. 1996, 281, 285–291. [Google Scholar] [CrossRef]
- Faber, E.J.; van den Haak, M.J.; Kamerling, J.P.; Vliegenthart, J.F.G. Structure of the Exopolysaccharide Produced by Streptococcus thermophilus S3. Carbohydr. Res. 2001, 331, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, K.; Suzuki, M.; Inukai, K.; Kuga, H.; Korenaga, H. Structural Study on a Sulfated Polysaccharide-Peptidoglycan Complex Produced by Arthrobacter sp. Biosci. Biotechnol. Biochem. 1998, 62, 2138–2144. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 267–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latge, J.-P. Galactofuranose Containing Molecules in Aspergillus fumigatus. Med. Mycol. 2009, 47, 104–109. [Google Scholar] [CrossRef] [Green Version]
- Latge, J.-P. Aspergillus fumigatus and Aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [CrossRef] [Green Version]
- Oka, T. Biosynthesis of Galactomannans Found in Filamentous Fungi Belonging to Pezizomycotina. Biosci. Biotechnol. Biochem. 2018, 82, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Matveev, A.L.; Krylov, V.B.; Emelyanova, L.A.; Solovev, A.S.; Khlusevich, Y.A.; Baykov, I.K.; Fontaine, T.; Latgé, J.-P.; Tikunova, N.V.; Nifantiev, N.E. Novel Mouse Monoclonal Antibodies Specifically Recognize Aspergillus fumigatus Galactomannan. PLOS ONE 2018, 13, e0193938. [Google Scholar] [CrossRef] [Green Version]
- Jansson, P.-E.; Lindberg, B. Structural Studies of Varianose. Carbohydr. Res. 1980, 82, 97–102. [Google Scholar] [CrossRef]
- de Lederkremer, R.M.; Casal, O.L.; Alves, M.J.M.; Colli, W. Evidence for the Presence of D-Galactofuranose in the Lipopeptidophosphoglycan from Trypanosoma cruzi. FEBS Lett. 1980, 116, 25–29. [Google Scholar] [CrossRef] [Green Version]
- de Lederkremer, R.M.; Colli, W. Galactofuranose-Containing Glycoconjugates in Trypanosomatids. Glycobiology 1995, 5, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Cabezas, Y.; Legentil, L.; Robert-Gangneux, F.; Daligault, F.; Belaz, S.; Nugier-Chauvin, C.; Tranchimand, S.; Tellier, C.; Gangneux, J.-P.; Ferrières, V. Leishmania Cell Wall as a Potent Target for Antiparasitic Drugs. A Focus on the Glycoconjugates. Org. Biomol. Chem. 2015, 13, 8393–8404. [Google Scholar] [CrossRef]
- Oppenheimer, M.; Valenciano, A.L.; Sobrado, P. Biosynthesis of Galactofuranose in Kinetoplastids: Novel Therapeutic Targets for Treating Leishmaniasis and Chagas’ Disease. Enzyme Res. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, M.A.; Roberts, K. Methylation Analysis of Cell Wall Glycoproteins and Glycopeptides from Chlamydomonas reinhardii. Phytochemistry 1981, 20, 25–28. [Google Scholar] [CrossRef]
- Igarashi, T.; Satake, M.; Yasumoto, T. Structures and Partial Stereochemical Assignments for Prymnesin-1 and Prymnesin-2: Potent Hemolytic and Ichthyotoxic Glycosides Isolated from the Red Tide Alga Prymnesium Parvum. J. Am. Chem. Soc. 1999, 121, 8499–8511. [Google Scholar] [CrossRef]
- Pettit, G.R.; Xu, J.; Gingrich, D.E.; Williams, M.D.; Doubek, D.L.; Chapuis, J.-C.; Schmidt, J.M. Antineoplastic Agents. Part 395.1 Isolation and Structure of Agelagalastatin from the Papua New Guinea Marine Sponge Agelas sp. Chem. Commun. 1999, 10, 915–916. [Google Scholar] [CrossRef]
- Novelli, J.F.; Chaudhary, K.; Canovas, J.; Benner, J.S.; Madinger, C.L.; Kelly, P.; Hodgkin, J.; Carlow, C.K.S. Characterization of the Caenorhabditis elegans UDP-Galactopyranose Mutase Homolog Glf-1 Reveals an Essential Role for Galactofuranose Metabolism in Nematode Surface Coat Synthesis. Dev. Biol. 2009, 335, 340–355. [Google Scholar] [CrossRef] [Green Version]
- Houseknecht, J.B.; Lowary, T.L. Chemistry and Biology of Arabinofuranosyl- and Galactofuranosyl-Containing Polysaccharides. Curr. Opin. Chem. Biol. 2001, 5, 677–682. [Google Scholar] [CrossRef]
- Nassau, P.M.; Martin, S.L.; Brown, R.E.; Weston, A.; Monsey, D.; McNeil, M.R.; Duncan, K. Galactofuranose Biosynthesis in Escherichia coli K-12: Identification and Cloning of UDP-Galactopyranose Mutase. J. Bacteriol. 1996, 178, 1047–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanner, J.J.; Boechi, L.; Andrew McCammon, J.; Sobrado, P. Structure, Mechanism, and Dynamics of UDP-Galactopyranose Mutase. Arch. Biochem. Biophys. 2014, 544, 128–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kizjakina, K.; Tanner, J.J.; Sobrado, P. Targeting UDP-Galactopyranose Mutases from Eukaryotic Human Pathogens. Curr. Pharm. Des. 2013, 19, 2561–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, M.R.; Lowary, T.L. Chemistry and Biology of Galactofuranose-Containing Polysaccharides. ChemBioChem 2009, 10, 1920–1938. [Google Scholar] [CrossRef] [PubMed]
- Poulin, M.B.; Zhou, R.; Lowary, T.L. Synthetic UDP-Galactofuranose Analogs Reveal Critical Enzyme–Substrate Interactions in GlfT2-Catalyzed Mycobacterial Galactan Assembly. Org. Biomol. Chem. 2012, 10, 4074. [Google Scholar] [CrossRef]
- Wheatley, R.W.; Zheng, R.B.; Richards, M.R.; Lowary, T.L.; Ng, K.K.S. Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan. J. Biol. Chem. 2012, 287, 28132–28143. [Google Scholar] [CrossRef] [Green Version]
- Wing, C.; Errey, J.C.; Mukhopadhyay, B.; Blanchard, J.S.; Field, R.A. Expression and Initial Characterization of WbbI, a Putative d-Galf: α-d-Glc β-1,6-Galactofuranosyltransferase from Escherichia coli K-12. Org Biomol Chem 2006, 4, 3945–3950. [Google Scholar] [CrossRef]
- Guan, S.; Clarke, A.J.; Whitfield, C. Functional Analysis of the Galactosyltransferases Required for Biosynthesis of D-Galactan I, a Component of the Lipopolysaccharide O1 Antigen of Klebsiella pneumoniae. J. Bacteriol. 2001, 183, 3318–3327. [Google Scholar] [CrossRef] [Green Version]
- Ati, J.; Colas, C.; Lafite, P.; Sweeney, R.P.; Zheng, R.B.; Lowary, T.L.; Daniellou, R. The LPG1x Family from Leishmania major Is Constituted of Rare Eukaryotic Galactofuranosyltransferases with Unprecedented Catalytic Properties. Sci. Rep. 2018, 8, 17566. [Google Scholar] [CrossRef]
- Komachi, Y.; Hatakeyama, S.; Motomatsu, H.; Futagami, T.; Kizjakina, K.; Sobrado, P.; Ekino, K.; Takegawa, K.; Goto, M.; Nomura, Y.; et al. GfsA Encodes a Novel Galactofuranosyltransferase Involved in Biosynthesis of Galactofuranose Antigen of O-Glycan in Aspergillus nidulans and Aspergillus fumigatus. Mol. Microbiol. 2013, 90, 1054–1073. [Google Scholar] [CrossRef] [Green Version]
- Katafuchi, Y.; Li, Q.; Tanaka, Y.; Shinozuka, S.; Kawamitsu, Y.; Izumi, M.; Ekino, K.; Mizuki, K.; Takegawa, K.; Shibata, N.; et al. GfsA Is a Β1,5-Galactofuranosyltransferase Involved in the Biosynthesis of the Galactofuran Side Chain of Fungal-Type Galactomannan in Aspergillus fumigatus. Glycobiology 2017, 27, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Mariño, K.; Marino, C.; Lima, C.; Baldoni, L.; de Lederkremer, R.M. The First Chemical Synthesis of UDP[6 -3H]-α-d-Galactofuranose. Eur. J. Org. Chem. 2005, 2005, 2958–2964. [Google Scholar] [CrossRef]
- Rietschel-Berst, M.; Jentoft, N.H.; Rick, P.D.; Pletcher, C.; Fang, F.; Gander, J.E. Extracellular Exo-Beta-Galactofuranosidase from Penicillium charlesii: Isolation, Purification, and Properties. J. Biol. Chem. 1977, 252, 3219–3226. [Google Scholar] [PubMed]
- Daley, L.S.; Strobel, G.A. β-Galactofuranosidase Activity in Helminthosporium sacchari and Its Relationship to the Production of Helminthosporoside. Plant Sci. Lett. 1983, 30, 145–154. [Google Scholar] [CrossRef]
- Livingston, R.S.; Scheffer, R.P. Conversion of Helminthosporium sacchari Toxin to Toxoids by β-Galactofuranosidase from Helminthosporium. Plant Physiol. 1983, 72, 530–534. [Google Scholar] [CrossRef] [Green Version]
- Lugt, A.W.V.B.-V.D.; Kamphuis, H.J.; Ruiter, G.A.D.; Mischnick, P.; Boom, J.H.V.; Rombouts, F.M. New Structural Features of the Antigenic Extracellular Polysaccharides of Penicillium and Aspergillus Species Revealed with Exo-Beta-d-Galactofuranosidase. J. Bacteriol. 1992, 174, 6096–6102. [Google Scholar] [CrossRef] [Green Version]
- Cousin, M.A.; Notermans, S.; Hoogerhout, P.; Van Boom, J.H. Detection of Beta-Galactofuranosidase Production by Penicillium and Aspergillus Species Using 4-Nitrophenyl Beta-d-Galactofuranoside. J. Appl. Bacteriol. 1989, 66, 311–317. [Google Scholar] [CrossRef]
- Pletcher, C.H.; Lomar, P.D.; Gander, J.E. Factors Affecting the Accumulation of Exocellularexo-β-d-Galactofuranosidase and Other Enzymes From Penicillium charlesii. Exp. Mycol. 1981, 5, 133–139. [Google Scholar] [CrossRef]
- Miletti, L.C.; Marino, C.; Colli, W. Immobilized 4-Aminophenyl 1-Thio-b-d-Galactofuranoside as a Matrix for Affinity Purification of an Exo-b-d-Galactofuranosidase. Carbohydr. Res. 1999, 320, 176–182. [Google Scholar] [CrossRef]
- Tuekam, B.A.; Park, Y.-I.; Unkefer, C.J.; Gander, J.E. Relationship of Exo-β-d-Galactofuranosidase Kinetic Parameters to the Number of Phosphodiesters in Penicillium fellutanum Peptidophosphogalactomannan: Enzyme Purification and Kinetics of Glycopeptide and Galactofuran Chain Hydrolysis. Appl. Environ. Microbiol. 2001, 67, 4648–4656. [Google Scholar] [CrossRef] [Green Version]
- Wallis, G.L.F.; Hemming, F.W.; Peberdy, J.F. An Extracellular L-Galactofuranosidase from Aspergillus niger and Its Use as a Tool for Glycoconjugate Analysis. Biochim. Biophys. Acta 2001, 1525, 19–28. [Google Scholar] [CrossRef]
- Miletti, L.C.; Mariño, K.; Marino, C.; Colli, W.; Alves, M.J.M.; de Lederkremer, R.M. Evidence for Exo β-d-Galactofuranosidase in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2003, 127, 85–88. [Google Scholar] [CrossRef]
- Reyes, F.; Alfonso, C.; Martinez, M.-J.; Prieto, A.; Santamaria, F.; Leal, J.-A. Purification of a New Galactanase from Penicillium oxalicum Catalysing the Hydrolysis of β-(1→5)-Galactofuran Linkages. Biochem. J. 1992, 281, 657–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramli, N.; Fujinaga, M.; Tabuchi, M.; Takegawa, K.; Iwahara, S. Isolation and Characterization of a Novel Endo-β-Galactofuranosidase from Bacillus sp. Biosci. Biotechnol. Biochem. 1995, 59, 1856–1860. [Google Scholar] [CrossRef]
- Mennink-Kersten, M.A.S.H.; Ruegebrink, D.; Wasei, N.; Melchers, W.J.G.; Verweij, P.E. In Vitro Release by Aspergillus fumigatus of Galactofuranose Antigens, 1,3-β-d-Glucan, and DNA, Surrogate Markers Used for Diagnosis of Invasive Aspergillosis. J. Clin. Microbiol. 2006, 44, 1711–1718. [Google Scholar] [CrossRef] [Green Version]
- Mariño, K.; Lima, C.; Maldonado, S.; Marino, C.; de Lederkremer, R.M. Influence of Exo Beta-d-Galactofuranosidase Inhibitors in Cultures of Penicillium fellutanum and Modifications in Hyphal Cell Structure. Carbohydr. Res. 2002, 337, 891–897. [Google Scholar] [CrossRef]
- Dubourdieu, D.; Desplanques, C.; Villetaz, J.-C.; Ribereau-Gayon, P. Investigations of an Industrial β-d-Glucanase from Trichoderma harzianum. Carbohydr. Res. 1985, 144, 277–287. [Google Scholar] [CrossRef]
- Marino, C.; Mariño, K.; Miletti, L.; Manso Alves, M.J.; Colli, W.; de Lederkremer, R.M. 1-Thio-β-d-Galactofuranosides: Synthesis and Evaluation as β-d-Galactofuranosidase Inhibitors. Glycobiology 1998, 8, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Varela, O.; Marino, C.; de Lederkremer, R.M. Synthesis of p-Nitrophenyl β-d-Galactofuranoside. A Convenient Substrate for β-Galactofuranosidase. Carbohydr. Res. 1986, 155, 247–251. [Google Scholar] [CrossRef]
- Matsunaga, E.; Higuchi, Y.; Mori, K.; Tashiro, K.; Kuhara, S.; Takegawa, K. Draft Genome Sequence of Streptomyces sp. JHA19, a Strain That Possesses β-d-Galactofuranosidase Activity. Genome Announc. 2015, 3, e01171-15. [Google Scholar] [CrossRef] [Green Version]
- Matsunaga, E.; Higuchi, Y.; Mori, K.; Yairo, N.; Oka, T.; Shinozuka, S.; Tashiro, K.; Izumi, M.; Kuhara, S.; Takegawa, K. Identification and Characterization of a Novel Galactofuranose-Specific β-d-Galactofuranosidase from Streptomyces Species. PLOS ONE 2015, 10, e0137230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, E.; Higuchi, Y.; Mori, K.; Yairo, N.; Toyota, S.; Oka, T.; Tashiro, K.; Takegawa, K. Characterization of a PA14 Domain-Containing Galactofuranose-Specific β-d-Galactofuranosidase from Streptomyces sp. Biosci. Biotechnol. Biochem. 2017, 81, 1314–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, E.; Higuchi, Y.; Mori, K.; Tashiro, K.; Takegawa, K. Draft Genome Sequence of Streptomyces sp. JHA26, a Strain That Harbors a PA14 Domain Containing β-d-Galactofuranosidase. Genome Announc. 2017, 5, e00190-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seničar, M.; Legentil, L.; Ferrières, V.; Eliseeva, S.V.; Petoud, S.; Takegawa, K.; Lafite, P.; Daniellou, R. Galactofuranosidase from JHA 19 Streptomyces sp.: Subcloning and Biochemical Characterization. Carbohydr. Res. 2019, 480, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Gagneux, P. Evolution of Carbohydrate Antigens—Microbial Forces Shaping Host Glycomes? Glycobiology 2007, 17, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Heesemann, L.; Kotz, A.; Echtenacher, B.; Broniszewska, M.; Routier, F.; Hoffmann, P.; Ebel, F. Studies on Galactofuranose-Containing Glycostructures of the Pathogenic Mold Aspergillus fumigatus. Int. J. Med. Microbiol. 2011, 301, 523–530. [Google Scholar] [CrossRef]
- Stynen, D.; Sarfati, J.; Goris, A.; Prévost, M.C.; Lesourd, M.; Kamphuis, H.; Darras, V.; Latgé, J.P. Rat Monoclonal Antibodies against Aspergillus Galactomannan. Infect. Immun. 1992, 60, 2237–2245. [Google Scholar] [CrossRef] [Green Version]
- Verdaguer, V.; Walsh, T.J.; Hope, W.; Cortez, K.J. Galactomannan Antigen Detection in the Diagnosis of Invasive Aspergillosis. Expert Rev. Mol. Diagn. 2007, 7, 21–32. [Google Scholar] [CrossRef]
- Marino, C.; Rinflerch, A.; de Lederkremer, R.M. Galactofuranose Antigens, a Target for Diagnosis of Fungal Infections in Humans. Future Sci. OA 2017, 3, FSO199. [Google Scholar] [CrossRef]
- Rolle, A.-M.; Hasenberg, M.; Thornton, C.R.; Solouk-Saran, D.; Männ, L.; Weski, J.; Maurer, A.; Fischer, E.; Spycher, P.R.; Schibli, R.; et al. ImmunoPET/MR Imaging Allows Specific Detection of Aspergillus fumigatus Lung Infection In Vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 1026–1033. [Google Scholar] [CrossRef] [Green Version]
- Davies, G.; Rolle, A.-M.; Maurer, A.; Spycher, P.R.; Schillinger, C.; Solouk-Saran, D.; Hasenberg, M.; Weski, J.; Fonslet, J.; Dubois, A.; et al. Towards Translational ImmunoPET/MR Imaging of Invasive Pulmonary Aspergillosis: The Humanised Monoclonal Antibody JF5 Detects Aspergillus Lung Infections In Vivo. Theranostics 2017, 7, 3398–3414. [Google Scholar] [CrossRef] [PubMed]








| Enzyme | Species | Substrate | pH | T (°C) | M (kDa) | KM (mM) † | Year | Reference |
|---|---|---|---|---|---|---|---|---|
| extracellular exo-β-d-Galf-ase | Penicillium charlesii (fungus) | pPGM a | 4 | 47 | – | – | 1977 | [46] |
| β-Galf-ase | Helminthosporium sacchari (fungus) | 1-O-methyl-β-Galf | 4.2 & 5.2 | 38 | – | – | 1983 | [47] |
| β-d-Galf-ase | H. sacchari (fungus) | HS toxin b | 4.6 | 37 | – | – | 1983 | [48] |
| extracellular β-d-Galf-ase | Penicillium spp. | pNP-β-d-Galf | 5 | 30 | – | – | 1989 | [50] |
| Aspergillus spp. (fungi) | ||||||||
| endo-β-Galf-ase | Penicillium oxalicum (fungus) | β-(1→5)-galactofuran | 5 | 37 | 77 | – | 1992 | [56] |
| exo-β-d-Galf-ase | Trichoderma harzianum (fungus) | EPS c | 4–4.5 | 35–40 | 35 | – | 1992 | [49] |
| endo-β-Galf-ase | Bacillus sp. (bacteria) | __ | 6 | 37 | 67 | – | 1995 | [57] |
| exo-β-d-Galf-ase | Penicillium fellutanum (fungus) | pNP-β-d-Galf | 3–6 | 37 | 70 | 0.3 | 1999 | [52] |
| extracellular β-Galf-ase | Aspergillus niger (fungus) | pNP-β-d-Galf | 3–4 | 37 | 90 | 4 | 2001 | [54] |
| exo-β-d-Galf-ase | P. fellutanum (fungus) | 1-O-methyl-β-Galf | 4–4.5 | 40 | 70 | 2.6 | 2001 | [53] |
| exo-β-d-Galf-ase | Trypanosoma cruzi (protozoa) | LPPG d | – | – | 55 | – | 2003 | [55] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seničar, M.; Lafite, P.; Eliseeva, S.V.; Petoud, S.; Landemarre, L.; Daniellou, R. Galactofuranose-Related Enzymes: Challenges and Hopes. Int. J. Mol. Sci. 2020, 21, 3465. https://doi.org/10.3390/ijms21103465
Seničar M, Lafite P, Eliseeva SV, Petoud S, Landemarre L, Daniellou R. Galactofuranose-Related Enzymes: Challenges and Hopes. International Journal of Molecular Sciences. 2020; 21(10):3465. https://doi.org/10.3390/ijms21103465
Chicago/Turabian StyleSeničar, Mateja, Pierre Lafite, Svetlana V. Eliseeva, Stéphane Petoud, Ludovic Landemarre, and Richard Daniellou. 2020. "Galactofuranose-Related Enzymes: Challenges and Hopes" International Journal of Molecular Sciences 21, no. 10: 3465. https://doi.org/10.3390/ijms21103465