Stress Granule Formation Attenuates RACK1-Mediated Apoptotic Cell Death Induced by Morusin
Abstract
:1. Introduction
2. Results
2.1. Morusin Induces Stress Granule Formation
2.2. PKR Activation Is Required for the Induction of Stress Granules by Morusin
2.3. Morusin-Induced Stress Granules Enhance Cell Survival by Inhibition of Cell Death
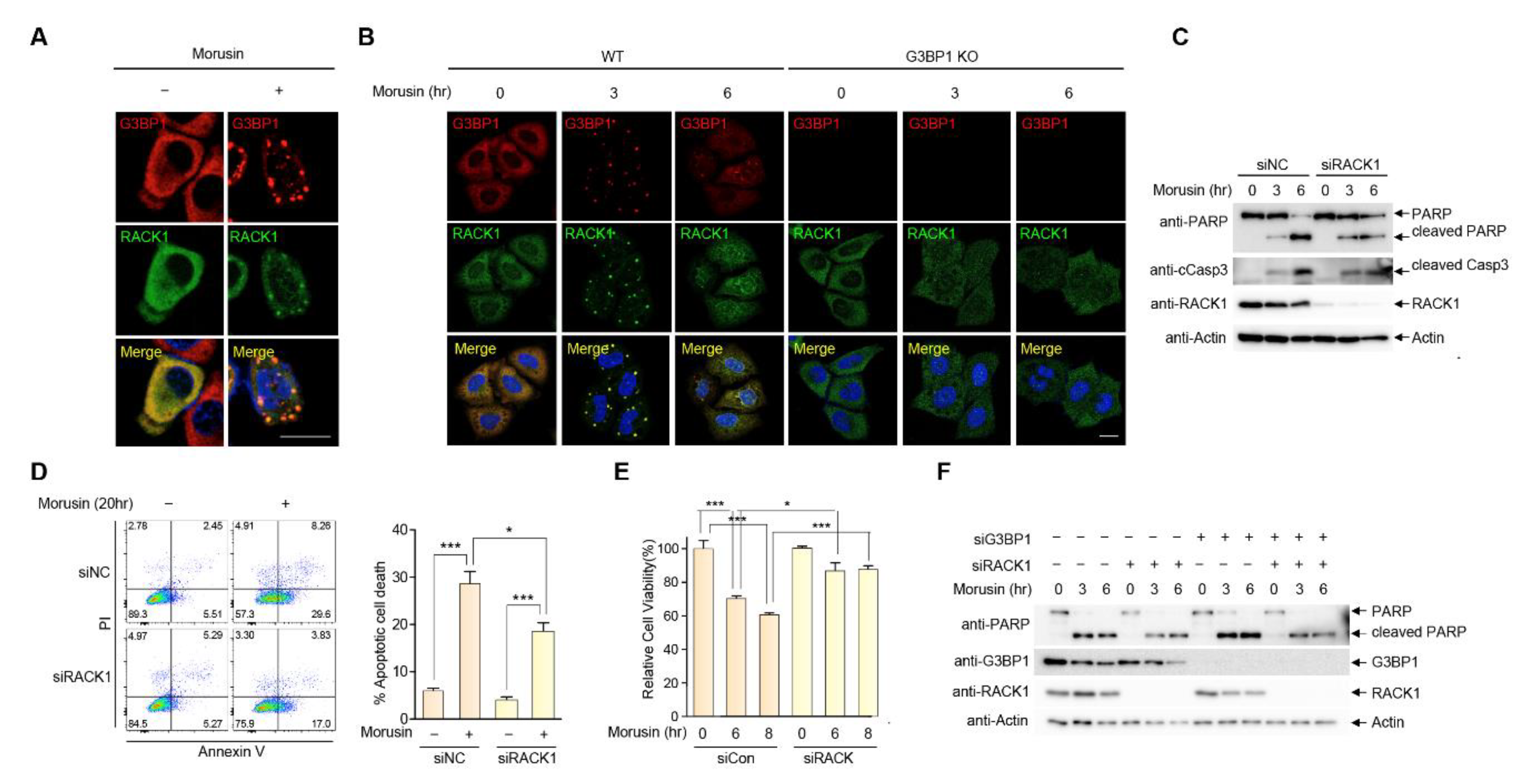
2.4. RACK1 Sequestration at Stress Granules Inhibits Apoptosis
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Antibodies and Reagents
4.3. DNA and siRNA Transfection
4.4. Generation of G3BP1 KO Cell Line
4.5. Immunocytochemistry
4.6. Immunoblot Analysis
4.7. Cell Viability Analysis
4.8. Annexin V-FITC/Propidium Iodide Double Staining
4.9. Colony Formation Assay
4.10. Antibody Array
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SG | Stress granule |
| PKR | Protein kinase R |
| RACK1 | Receptor for activated C kinase-1 |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
References
- Buchan, J.R.; Parker, R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [Green Version]
- Wallace, E.W.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tourrière, H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003, 160, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.; Dezi, V.; Pizzinga, M.; Willis, A.E. Post-transcriptional control of gene expression following stress: The role of RNA-binding proteins. Biochem. Soc. Trans. 2017, 45, 1007–1014. [Google Scholar] [CrossRef]
- Aulas, A.; Fay, M.M.; Lyons, S.M.; Achorn, C.A.; Kedersha, N.; Anderson, P.; Ivanov, P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017, 130, 927–937. [Google Scholar] [CrossRef] [Green Version]
- Solomon, S.; Xu, Y.; Wang, B.; David, M.D.; Schubert, P.; Kennedy, D.; Schrader, J.W. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 2007, 27, 2324–2342. [Google Scholar] [CrossRef] [Green Version]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [Green Version]
- Tsai, W.C.; Gayatri, S.; Reineke, L.C.; Sbardella, G.; Bedford, M.T.; Lloyd, R.E. Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J. Biol. Chem. 2016, 291, 22671–22685. [Google Scholar] [CrossRef] [Green Version]
- Chitiprolu, M.; Jagow, C.; Tremblay, V.; Bondy-Chorney, E.; Paris, G.; Savard, A.; Palidwor, G.; Barry, F.A.; Zinman, L.; Keith, J.; et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat. Commun. 2018, 9, 2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Maxwell, B.A.; Joo, J.H.; Gwon, Y.; Messing, J.; Mishra, A.; Shaw, T.I.; Ward, A.L.; Quan, H.; Sakurada, S.M.; et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol. Cell 2019, 74, 742–757. [Google Scholar] [CrossRef]
- Aditi; Mason, A.C.; Sharma, M.; Dawson, T.R.; Wente, S.R. MAPK- and glycogen synthase kinase 3-mediated phosphorylation regulates the DEAD-box protein modulator Gle1 for control of stress granule dynamics. J. Biol. Chem. 2019, 294, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reineke, L.C.; Tsai, W.C.; Jain, A.; Kaelber, J.T.; Jung, S.Y.; Lloyd, R.E. Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, J.Y.; Dyakov, B.J.A.; Zhang, J.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.C. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef]
- Thedieck, K.; Holzwarth, B.; Prentzell, M.T.; Boehlke, C.; Kläsener, K.; Ruf, S.; Sonntag, A.G.; Maerz, L.; Grellscheid, S.N.; Kremmer, E.; et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 2013, 154, 859–874. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.J.; Back, S.H.; Kim, V.; Ryu, I.; Jang, S.K. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 2005, 25, 2450–2462. [Google Scholar] [CrossRef] [Green Version]
- Arimoto, K.; Fukuda, H.; Imajoh-Ohmi, S.; Saito, H.; Takekawa, M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008, 10, 1324–1332. [Google Scholar] [CrossRef]
- Yoneyama, M.; Jogi, M.; Onomoto, K. Regulation of antiviral innate immune signaling by stress-induced RNA granules. J. Biochem. 2016, 159, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Onomoto, K.; Yoneyama, M.; Fung, G.; Kato, H.; Fujita, T. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014, 35, 420–428. [Google Scholar] [CrossRef]
- Wolozin, B.; Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666. [Google Scholar] [CrossRef]
- Anderson, P.; Kedersha, N.; Ivanov, P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 2015, 1849, 861–870. [Google Scholar] [CrossRef] [Green Version]
- Adjibade, P.; St-Sauveur, V.G.; Quevillon Huberdeau, M.; Fournier, M.J.; Savard, A.; Coudert, L.; Khandjian, E.W.; Mazroui, R. Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget 2015, 6, 43927–43943. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, C.; Isensee, J.; Hucho, T.; Lehrach, H.; Krobitsch, S. 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Res. 2014, 42, 6436–6447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somasekharan, S.P.; El-Naggar, A.; Leprivier, G.; Cheng, H.; Hajee, S.; Grunewald, T.G.; Zhang, F.; Ng, T.; Delattre, O.; Evdokimova, V.; et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015, 208, 913–929. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Fukai, T.; Matsumoto, J.; Ohmori, T. Constituents of the cultivated mulberry tree. Planta Med. 1982, 46, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Ryu, J.; Park, S.H.; Woo, E.R.; Kim, A.R.; Lee, S.K.; Kim, Y.S.; Kim, J.O.; Hong, J.H.; Lee, C.J. Effects of Morus alba L. and Natural Products Including Morusin on In Vivo Secretion and In Vitro Production of Airway MUC5AC Mucin. Tuberc. Respir. Dis. 2014, 77, 65–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singab, A.N.; El-Beshbishy, H.A.; Yonekawa, M.; Nomura, T.; Fukai, T. Hypoglycemic effect of Egyptian Morus alba root bark extract: Effect on diabetes and lipid peroxidation of streptozotocin-induced diabetic rats. J. Ethnopharmacol. 2005, 100, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lyu da, H.; Koo, U.; Nam, K.W.; Hong, S.S.; Kim, K.O.; Kim, K.H.; Lee, D.; Mar, W. Protection of prenylated flavonoids from Mori Cortex Radicis (Moraceae) against nitric oxide-induced cell death in neuroblastoma SH-SY5Y cells. Arch. Pharmacal Res. 2012, 35, 163–170. [Google Scholar] [CrossRef]
- Park, H.J.; Min, T.R.; Chi, G.Y.; Choi, Y.H.; Park, S.H. Induction of apoptosis by morusin in human non-small cell lung cancer cells by suppression of EGFR/STAT3 activation. Biochem. Biophys. Res. Commun. 2018, 505, 194–200. [Google Scholar] [CrossRef]
- Li, H.; Wang, Q.; Dong, L.; Liu, C.; Sun, Z.; Gao, L.; Wang, X. Morusin suppresses breast cancer cell growth in vitro and in vivo through C/EBPβ and PPARγ mediated lipoapoptosis. J. Exp. Clin. Cancer Res. CR 2015, 34, 137R. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Kim, E.O.; Kim, S.H.; Lee, J.H.; Ahn, K.S.; Yun, M.; Lee, S.G. Morusin induces apoptosis by regulating expression of Bax and Survivin in human breast cancer cells. Oncol. Lett. 2017, 13, 4558–4562. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Li, R.; Zhao, X.; Ma, C.; Lv, X.; Liu, L.; Liu, P. Morusin induces paraptosis-like cell death through mitochondrial calcium overload and dysfunction in epithelial ovarian cancer. Chem. Biol. Interact. 2018, 283, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, D.; Mao, J.; Ke, X.X.; Zhang, R.; Yin, C.; Gao, N.; Cui, H. Morusin inhibits cell proliferation and tumor growth by down-regulating c-Myc in human gastric cancer. Oncotarget 2017, 8, 57187–57200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.L.; Lv, Y.; Wang, S.; Zhang, Y.Q. Morusin suppresses A549 cell migration and induces cell apoptosis by downregulating the expression of COX-2 and VEGF genes. Oncol. Rep. 2018, 40, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Luo, J.; Luo, X.; Jia, W.; Fang, Z.; Yi, S.; Li, L. Morusin exerts anti-cancer activity in renal cell carcinoma by disturbing MAPK signaling pathways. Ann. Transl. Med. 2020, 8, 327. [Google Scholar] [CrossRef]
- Lim, S.L.; Park, S.Y.; Kang, S.; Park, D.; Kim, S.H.; Um, J.Y.; Jang, H.J.; Lee, J.H.; Jeong, C.H.; Jang, J.H.; et al. Morusin induces cell death through inactivating STAT3 signaling in prostate cancer cells. Am. J. Cancer Res. 2015, 5, 289–299. [Google Scholar]
- Kim, C.; Kim, J.H.; Oh, E.Y.; Nam, D.; Lee, S.G.; Lee, J.; Kim, S.H.; Shim, B.S.; Ahn, K.S. Blockage of STAT3 Signaling Pathway by Morusin Induces Apoptosis and Inhibits Invasion in Human Pancreatic Tumor Cells. Pancreas 2016, 45, 409–419. [Google Scholar] [CrossRef]
- Lin, W.L.; Lai, D.Y.; Lee, Y.J.; Chen, N.F.; Tseng, T.H. Antitumor progression potential of morusin suppressing STAT3 and NFκB in human hepatoma SK-Hep1 cells. Toxicol. Lett. 2015, 232, 490–498. [Google Scholar] [CrossRef]
- Gao, L.; Wang, L.; Sun, Z.; Li, H.; Wang, Q.; Yi, C.; Wang, X. Morusin shows potent antitumor activity for human hepatocellular carcinoma in vitro and in vivo through apoptosis induction and angiogenesis inhibition. Drug Des. Dev. Ther. 2017, 11, 1789–1802. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.W.; Na, W.; Choi, M.; Kang, S.J.; Lee, S.G.; Choi, C.Y. Autophagy inhibits cell death induced by the anti-cancer drug morusin. Am. J. Cancer Res. 2017, 7, 518–530. [Google Scholar] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L. Stress Granules and Virus Replication. Future Virol. 2011, 6, 1329–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheuner, D.; Song, B.; McEwen, E.; Liu, C.; Laybutt, R.; Gillespie, P.; Saunders, T.; Bonner-Weir, S.; Kaufman, R.J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 2001, 7, 1165–1176. [Google Scholar] [CrossRef]
- Buchan, J.R.; Kolaitis, R.M.; Taylor, J.P.; Parker, R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013, 153, 1461–1474. [Google Scholar] [CrossRef] [Green Version]
- Monahan, Z.; Shewmaker, F.; Pandey, U.B. Stress granules at the intersection of autophagy and ALS. Brain Res. 2016, 1649, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Sfakianos, A.P.; Mellor, L.E.; Pang, Y.F.; Kritsiligkou, P.; Needs, H.; Abou-Hamdan, H.; Désaubry, L.; Poulin, G.B.; Ashe, M.P.; Whitmarsh, A.J. The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 2018, 25, 1766–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heberle, A.M.; Razquin Navas, P.; Langelaar-Makkinje, M.; Kasack, K.; Sadik, A.; Faessler, E.; Hahn, U.; Marx-Stoelting, P.; Opitz, C.A.; Sers, C.; et al. The PI3K and MAPK/p38 pathways control stress granule assembly in a hierarchical manner. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef]
- Billmann, M.; Chaudhary, V.; ElMaghraby, M.F.; Fischer, B.; Boutros, M. Widespread Rewiring of Genetic Networks upon Cancer Signaling Pathway Activation. Cell Syst. 2018, 6, 52–64. [Google Scholar] [CrossRef]
- Gao, X.; Jiang, L.; Gong, Y.; Chen, X.; Ying, M.; Zhu, H.; He, Q.; Yang, B.; Cao, J. Stress granule: A promising target for cancer treatment. Br. J. Pharmacol. 2019, 176, 4421–4433. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.R.; Ron, D.; Kiely, P.A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal. CCS 2011, 9, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza, M.M.; Bittar, M.; Cechinel-Filho, V.; Yunes, R.A.; Messana, I.; Delle Monache, F.; Ferrari, F. Antinociceptive properties of morusin, a prenylflavonoid isolated from Morus nigra root bark. Z. Fur Nat. C J. Biosci. 2000, 55, 256–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abd El-Mawla, A.M.; Mohamed, K.M.; Mostafa, A.M. Induction of Biologically Active Flavonoids in Cell Cultures of Morus nigra and Testing their Hypoglycemic Efficacy. Sci. Pharm. 2011, 79, 951–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, D.; Liao, S.; Wang, W.; Mu, L.; Li, E.; Shen, W.; Liu, F.; Zou, Y. Destruction of the cell membrane and inhibition of cell phosphatidic acid biosynthesis in Staphylococcus aureus: An explanation for the antibacterial mechanism of morusin. Food Funct. 2019, 10, 6438–6446. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.-J.; Choi, D.W.; Cho, S.W.; Han, J.; Yang, S.; Choi, C.Y. Stress Granule Formation Attenuates RACK1-Mediated Apoptotic Cell Death Induced by Morusin. Int. J. Mol. Sci. 2020, 21, 5360. https://doi.org/10.3390/ijms21155360
Park Y-J, Choi DW, Cho SW, Han J, Yang S, Choi CY. Stress Granule Formation Attenuates RACK1-Mediated Apoptotic Cell Death Induced by Morusin. International Journal of Molecular Sciences. 2020; 21(15):5360. https://doi.org/10.3390/ijms21155360
Chicago/Turabian StylePark, Ye-Jin, Dong Wook Choi, Sang Woo Cho, Jaeseok Han, Siyoung Yang, and Cheol Yong Choi. 2020. "Stress Granule Formation Attenuates RACK1-Mediated Apoptotic Cell Death Induced by Morusin" International Journal of Molecular Sciences 21, no. 15: 5360. https://doi.org/10.3390/ijms21155360




