Combined Metabolome and Transcriptome Analyses Reveal the Effects of Mycorrhizal Fungus Ceratobasidium sp. AR2 on the Flavonoid Accumulation in Anoectochilus roxburghii during Different Growth Stages
Abstract
:1. Introduction
2. Results
2.1. Detecting Mycorrhizal Fungus Colonization in A. roxburghii
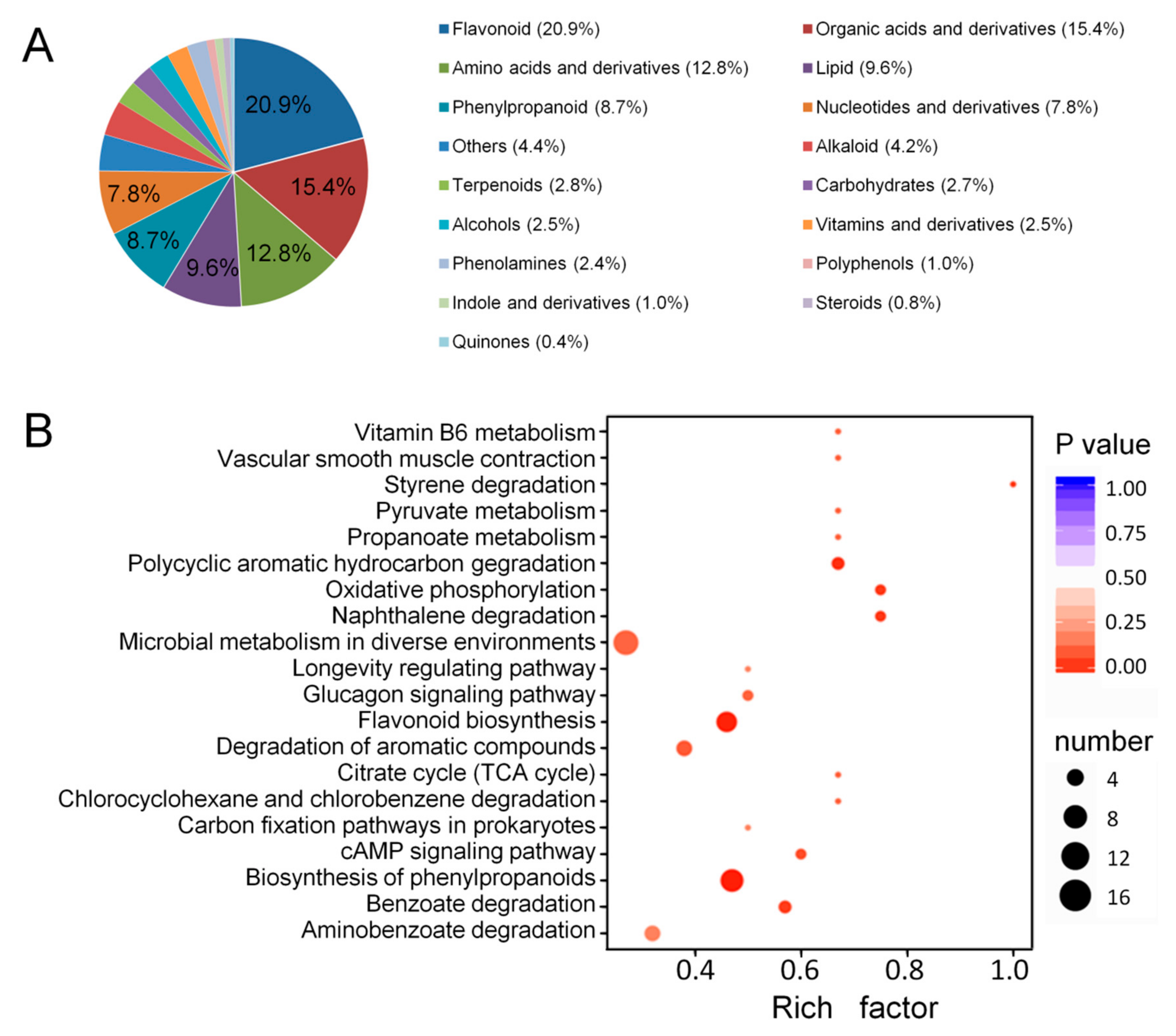
2.2. Identification of Metabolites
2.3. Identification of Differentially Accumulated Metabolites and Differentially Accumulated Flavonoids
2.4. Transcriptome Profiles of Mycorrhizal and Non-Mycorrhizal A. roxburghii
2.5. Association Analysis of the DAMs and DEGs
2.6. Dynamic Variations in Flavonoid DAMs
2.7. Dynamic Variations of Expression Levels of Flavonoid Biosynthetic Genes
3. Discussion
4. Materials and Methods
4.1. Plant and Mycorrhizal Fungus Materials
4.2. Symbiotic Cultures of A. roxburghii Plantlets
4.3. Histological Study
4.4. Sample Extraction and Metabolome Analysis
4.4.1. Sample Extraction
4.4.2. Liquid Chromatographic Mass Spectrometry Analysis
4.4.3. Metabolite Identification
4.5. Illumina Sequencing
4.5.1. RNA Extraction, cDNA Library Construction and Sequencing
4.5.2. De Novo Transcriptome Assembly and Annotation
4.6. Determination of the Flavonoid Contents During Different Growth Stages
4.6.1. Preparation of Standard Solutions
4.6.2. Preparation of Sample Solutions
4.6.3. Apparatus and Analytical Conditions
4.7. Expression of the Flavonoid Biosynthesis Related Genes During Different Growth Stages
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CE | collision energy |
| CUR | curtain gas |
| CXP | collision cell exit potential |
| DAMs | differentially accumulated metabolites |
| DEGs | differentially expressed genes |
| DP | de-clustering potential |
| EP | entrance potential |
| ESI-QTRAP-MS/MS | electrospray ionization-triple quadrupole-linear ion trap MS/MS |
| GC-MS | gas chromatography-mass spectrometer |
| GO | gene ontology |
| GS1 | ion source gas 1 |
| GS2 | ion source gas 2 |
| HPLC-MS/MS | high-performance liquid chromatography coupled with tandem mass spectrometry |
| IS | ion spray voltage |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LC-MS | liquid chromatograph-mass spectrometer |
| LIT | linear ion trap |
| LOD | limit of detection |
| LOQ | limit of quantification |
| NMR | nuclear magnetic resonance |
| OPLS-DA | orthogonal partial least squares discriminant |
| PCA | principal component analysis |
| PC1 | principal component 1 |
| PC2 | principal component 2 |
| PI | product ions |
| QC | quality control |
| QQQ | triple quadrupole |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| RNA-seq | RNA-sequencing |
| RSDs | relative standard deviations |
| TEM | temperature |
| TIC | total ion chromatography |
| UPLC-ESI-MS/MS | ultra-performance liquid chromatography-electrospray ionization-tandem mass spectrometry |
| VIP | variable importance in project |
| cps | count per second |
| MRM | multiple reaction monitoring |
| psi | pounds per square inch |
| NM | non-mycorrhizal A. roxburghii |
| M | mycorrhizal A. roxburghii |
| PAL | phenylalanine ammonia-lyase |
| C4H | cinnamate 4-hydroxylase |
| 4CL | 4-coumarate CoA ligase |
| CHS | chalcone synthase |
| CHI | chalcone isomerase |
| F3′H | flavonoid 3′-hydroxylase |
| FNS | flavone synthase |
| F3H | flavanone 3-hydroxylase |
| FLS | flavonol synthase |
| GT | flavonoid 3-O-glucosyltransferase |
| RT | rhamnosyltransferase |
References
- Li, S.; Wang, Z.; Shao, Q.; Fang, H.; Zhu, J.; Wu, X.; Zheng, B. Rapid detection of adulteration in Anoectochilus roxburghii by near-infrared spectroscopy coupled with chemometric methods. J. Food Sci. Technol. 2018, 55, 3518–3525. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, J.; Yeap, Z.Q.; Zhang, X.; Wu, S.; Ng, C.H.; Yam, M.F. Rapid authentication and identification of different types of A. roxburghii by Tri-step FT-IR spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 199, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.Y.; Su, M.H.; Chen, Q.X.; Chang, Q.; Wang, W.; Li, H.H. Antioxidant and hepatoprotective activities of polysaccharides from Anoectochilus roxburghii. Carbohydr. Polym. 2016, 153, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.C.; Yu, J.; Zhang, X.H.; Cheng, M.Z.; Yang, L.W.; Xu, J.Y. Antihyperglycemic and antioxidant activity of water extract from Anoectochilus roxburghii in experimental diabetes. Exp. Toxicol. Pathol. 2013, 65, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.; Duan, X.Y.; Zhang, L.; Shen, Y.B.; Hu, B.; Liu, A.P.; Chen, H.; Li, C.; Wu, W.J.; Shen, L.; et al. Antidiabetic activities of polysaccharides from Anoectochilus roxburghii and Anoectochilus formosanus in STZ-induced diabetic mice. Int. J. Biol. Macromol. 2018, 112, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.M.; Liu, Z.L.; Zhang, J.G.; Liu, Q.; Yi, L.T. The renal protective effects of Anoectochilus roxburghii polysaccharose on diabetic mice induced by high-fat diet and streptozotocin. J. Ethnopharmacol. 2016, 178, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.L.; Ye, Q.; Yang, S.L. Therapeutic effects of polysaccharides from Anoectochilus roxburghii on type II collagen-induced arthritis in rats. Int. J. Biol. Macromol. 2019, 122, 882–892. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, J.; Ruan, H.; Pi, H.; Wu, J. Antihyperglycemic activity of kinsenoside, a high yielding constituent from Anoectochilus roxburghii in streptozotocin diabetic rats. J. Ethnopharmacol. 2007, 114, 141–145. [Google Scholar] [CrossRef]
- Hsiao, H.B.; Wu, J.B.; Lin, H.; Lin, W.C. Kinsenoside isolated from Anoectochilus formosanus suppresses LPS-stimulated inflammatory reactions in macrophages and endotoxin shock in mice. Shock 2011, 35, 184–190. [Google Scholar] [CrossRef]
- Ye, S.Y.; Shao, Q.S.; Zhang, A.L. Anoectochilus roxburghii: A review of its phytochemistry, pharmacology, and clinical applications. J. Ethnopharmacol. 2017, 209, 184–202. [Google Scholar] [CrossRef]
- Dearnaley, J.D.W.; Martos, F.; Selosse, M.A. Orchid mycorrhizas: Molecular ecology, physiology, evolution and conservation aspects. In Fungal Association, 1st ed.; Hock, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 9, pp. 207–230. [Google Scholar]
- Dearnaley, J.D.; Cameron, D.D. Nitrogen transport in the orchid mycorrhizal symbiosis-further evidence for a mutualistic association. New Phytol. 2017, 213, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Jia, M.; Chen, L.; Zheng, C.J.; Rahman, K.; Han, T.; Qin, L.P. The regulatory mechanism of fungal elicitor-induced secondary metabolite biosynthesis in medical plants. Crit. Rev. Microbiol. 2017, 43, 238–261. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, G.; Li, B.; Guo, S.X. Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile Lindl. infected with mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Li, B.; Zhou, L.S.; Ding, G.; Li, B.; Guo, S.X. Molecular analysis of polysaccharide accumulation in Dendrobium nobile infected with the mycorrhizal fungus Mycena sp. RSC Adv. 2017, 7, 25872–25884. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.S.; Lv, Y.L.; Zhao, Y.; Guo, S.X. Promoting role of an endophyte on the growth and contents of kinsenosides and flavonoids of Anoectochilus formosanus hayata, a rare and threatened medicinal orchidaceae plant. J. Zhejiang Univ. Sci. B 2013, 14, 785–792. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.J.; Huang, Y.J.; Jin, J.H.; Shen, J.Y. Effect of cultivation substrate on growth and active component contents of Anoectochilus roxburghii from three different origins. China J. Chin. Mater. Med. 2019, 44, 2467–2471. [Google Scholar]
- Niu, H.; Xie, Z.M.; Gu, L.; Liang, Y.; Wei, K.H.; Wang, J.M.; Li, M.J.; Zhang, Z.Y. Effects of planting density and harvesting stages for Anoectochilus roxburghii planted under forest on its yield and quality. Mod. Chin. Med. 2018, 20, 837–865. [Google Scholar]
- Gan, J.J.; Mao, L.L.; Huang, R.L.; Jiang, H.Y.; Huang, X.Y.; Li, H. Effects of different cultivation methods on the growth and quality of Anoectochilus roxburghii. J. Agric. Sci. Technol. 2018, 20, 130–136. [Google Scholar]
- Wang, H.; Lin, Q.Q.; Hu, X.J.; Wu, G.H.; Xu, Q. Screening of Anoectochilus roxburghii growth-promoting endophytic fungi and their promoting mechanism. J. Fujian Norm. Univ. (Nat. Sci. Ed.) 2019, 35, 72–79. [Google Scholar]
- Deng, N.; Chang, E.; Li, M.H.; Ji, J.; Yao, X.M.; Bartish, I.V.; Liu, J.F.; Ma, J.; Chen, L.Z.; Jiang, Z.P.; et al. Transcriptome characterization of Gnetum parvifolium reveals candidate genes involved in important secondary metabolic pathways of flavonoids and stilbenoids. Front. Plant Sci. 2016, 7, 174. [Google Scholar] [CrossRef]
- Yue, J.Y.; Zhu, C.X.; Zhou, Y.; Niu, X.L.; Miao, M.; Tang, X.F.; Chen, F.D.; Zhao, W.P.; Liu, Y.S. Transcriptome analysis of differentially expressed unigenes involved in flavonoid biosynthesis during flower development of Chrysanthemum morifolium ‘Chu ju’. Sci. Rep. 2018, 8, 13414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, N.; Aoki, T.; Sato, S.; Nakamura, Y.; Tabata, S.; Ayabe, S. A cluster of genes encodes the two types of chalcone is omerase involved in the biosynthesis of general falavonoids and legume specific 5-deoxy (iso) flavonoids in Lotus japonicus. Plant Physiol. 2003, 131, 941–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nag, S.; Kumaria, S. In silico characterization and transcriptional modulation of phenylalanine ammonia lyase (PAL) by abiotic stresses in the medicinal orchid Vanda coerulea Griff. ex Lindl. Phytochemistry 2018, 156, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Yu, J.; Cai, Y.X.; Zhu, P.P.; Liu, C.Y.; Zhao, A.C.; Lü, R.H.; Li, M.J.; Xu, F.X.; Yu, M.D. Characterization and functional analysis of 4-coumarate: CoA ligase genes in mulberry. PLoS ONE 2016, 11, e0155814. [Google Scholar]
- Kim, J.; Woo, H.R.; Nam, H.G. Toward systems understanding of leaf senescence: An integrated multi-omics perspective on leaf senescence research. Mol. Plant 2016, 9, 813–825. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.Z.; Hu, Y.; Jiang, W.K.; Fang, L.; Guan, X.Y.; Chen, J.D.; Zhang, J.B.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.X.; Wang, W.; Liao, D.Q.; Wu, X.Y.; Li, X.E. UPLC-Q/TOF-MS-based serum metabolomics reveals hypoglycemic effects of Rehmannia glutinosa, Coptis chinensis and their combination on high-fat-diet-induced diabetes in KK-Ay Mice. Int. J. Mol. Sci. 2018, 19, 3984. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.L.; Zhang, J.X.; Chen, C.L.; Yang, J.Z.; Zhu, H.Y.; Liu, M.; Lv, F.B. Deep sequencing-based comparative transcriptional profiles of Cymbidium hybridum roots in response to mycorrhizal and non-mycorrhizal beneficial fungi. BMC Genom. 2014, 15, 747. [Google Scholar] [CrossRef] [Green Version]
- Sadre, R.; Magallanes-Lundback, M.; Pradhan, S.; Salim, V.; Mesberg, A.; Jones, A.D.; DellaPenna, D. Metabolite diversity in alkaloid biosynthesis: A multi-lane (diastereomer) highway for camptothecin synthesis in Camptotheca acuminate. Plant Cell 2016, 28, 1926–1944. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Luo, S.H.; Schmidt, A.; Wang, G.D.; Sun, G.L.; Grant, M.; Kuang, C.; Yang, M.J.; Jing, S.X.; Li, C.H.; et al. A geranylfarnesyl diphosphate synthase provides the precursor for sesterterpenoid (C25) formation in the glandular trichomes of the mint species Leucosceptrum canum. Plant Cell 2016, 28, 804–822. [Google Scholar] [CrossRef] [Green Version]
- Lou, Q.; Liu, Y.L.; Qi, Y.Y.; Jiao, S.Z.; Tian, F.F.; Jiang, L.; Wang, Y.J. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.; Cho, K.S.; Sohn, H.B.; Ha, I.J.; Hong, S.Y.; Lee, H.; Kim, Y.M.; Nam, M.H. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 2016, 67, 1519–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, D.; Zhou, R.; Wang, X.; Dossa, K.; Wang, L.; Zhang, Y.; Yu, J.; Gong, H.; Zhang, X.; et al. Transcriptome and metabolome analyses of two contrasting sesame genotypes reveal the crucial biological pathways involved in rapid adaptive response to salt stress. BMC Plant Biol. 2019, 19, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Jiang, C.; Ye, T.; Tan, D.X.; Reiter, R.J.; Zhang, H.; Liu, R.; Chan, Z. Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 2015, 66, 681–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broekgaarden, C.; Pelgrom, K.T.B.; Bucher, J.; van Dam, N.M.; Grosser, K.; Pieterse, C.M.J.; van Kaauwen, M.; Steenhuis, G.; Voorrips, R.E.; de Vos, M.; et al. Combining QTL mapping with transcriptome and metabolome profiling reveals a possible role for ABA signaling in resistance against the cabbage whitefly in cabbage. PLoS ONE 2018, 13, e0206103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, L.; Xie, L.; He, Y.; Luo, T.; Sheng, L.; Luo, Y.; Zeng, Y.; Xu, J.; Deng, X.; et al. Regulation of cuticle formation during fruit development and ripening in ‘Newhall’ navel orange (Citrus sinensis Osbeck) revealed by transcriptomic and metabolomic profiling. Plant Sci. 2016, 243, 131–144. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: San Diego, CA, USA, 2008; pp. 419–506. [Google Scholar]
- Harrison, M.J. Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 2005, 59, 19–42. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Timmers, T.; Bonfante, P.; Barker, D.G. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 2005, 17, 3489–3499. [Google Scholar] [CrossRef] [Green Version]
- Adolfsson, L.; Nziengui, H.; Abreu, I.N.; Šimura, J.; Beebo, A.; Herdean, A.; Aboalizadeh, J.; Široká, J.; Moritz, T.; Novák, O.; et al. Enhanced secondary-and hormone metabolism in leaves of arbuscular mycorrhizal Medicago truncatula. Plant Physiol. 2017, 175, 392–411. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.P.; Staehelin, C.; Vierheilig, H.; Wiemken, A.; Jabbouri, S.; Broughton, W.J.; Vogeli-Lange, R.; Boller, T. Rhizobial nodulation factors stimulate mycorrhizal colonization of nodulating and nonnodulating soybeans. Plant Physiol. 1995, 108, 1519–1525. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, Y.Y.; Guo, S.X. Effects of the mycorrhizal fungus Ceratobasidium sp. AR2 on growth and flavonoid accumulation in Anoectochilus roxburghii. PeerJ 2019, 7, e8346. [Google Scholar]
- Schliemann, W.; Ammer, C.; Strack, D. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 2008, 69, 112–146. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Hong, W.J.; Huang, J.X.; Tang, G.D. Impact of inoculation with mycorrhizal fungi in vitro on growth and resistant enzymes of Rhododendron moulmainense. Southwest China J. Argric. Sci. 2017, 30, 2687–2692. [Google Scholar]
- Xu, C.; Zhang, H.Y.; Liu, G.H.; Yang, H.T.; Xi, G.J. The effect and disease-resistant mechanism of mycorrhizal fungus on Dendrobium officinale Kimura et Migo. J. West China Sci. 2017, 46, 1–5. [Google Scholar]
- Han, Y.Y.; Ming, F.; Wang, W. Molecular evolution and functional specialization of chalcone synthase superfamily from Phalaenopsis orchid. Genetica 2006, 128, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Dixon, R.A. Spatial patterns of expression of flavonoid/isoflavonoid pathway genes during interactions between roots of Medicago truncatula and the mycorrhizal fungus Glomus versiforme. Plant J. 1994, 6, 9–20. [Google Scholar] [CrossRef]
- Xie, W.; Hao, Z.P.; Zhou, X.F.; Jiang, X.L.; Xu, L.J.; Wu, S.L.; Zhao, A.H.; Zhang, X.; Chen, B.D. Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza 2018, 28, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Wei, K.; Li, H.L.; Wang, L.Y.; Li, R.; Pang, D.D.; Cheng, H. Identification of key genes involved in catechin metabolism in tea seedlings based on transcriptomic and HPLC analysis. Plant Physiol. Biochem. 2018, 133, 107–115. [Google Scholar] [CrossRef]
- Park, S.; Kim, D.H.; Lee, J.Y.; Ha, S.H.; Lim, S.H. Comparative analysis of two flavonol synthases from different-colored onions provides insights into flavonoid biosynthesis. J. Agric. Food Chem. 2017, 65, 5287–5298. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| NM1 | 61,226,728 | 61,071,914 | 9.09 | 0.017 | 97.79 | 93.61 | 48.19 |
| NM2 | 60,542,772 | 60,425,910 | 8.99 | 0.016 | 98.11 | 94.42 | 47.93 |
| NM3 | 67,559,786 | 67,410,292 | 10.05 | 0.016 | 98.07 | 94.28 | 48.43 |
| M1 | 55,632,192 | 55,492,010 | 8.28 | 0.016 | 98.02 | 94.20 | 49.06 |
| M2 | 65,007,376 | 64,859,884 | 9.68 | 0.016 | 98.19 | 94.58 | 48.10 |
| M3 | 67,125,158 | 66,965,132 | 9.99 | 0.016 | 98.09 | 94.34 | 48.23 |
| No. | Name | Precursor Ion (m/z) | Product Ions (m/z) | DP (V) | CE (V) | EP (V) | CXP (V) | IS (V) | Ionization Mode | Retention Time (min) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PI q | PI i | ||||||||||
| 1 | nobiletin | 402.9 | 373.1 | 388.1 | 20 | 33 | 10 | 13 | 5500 | ESI+ | 4.56 |
| 2 | narcissin | 624.9 | 316.7 | 479.6 | 50 | 25 | 10 | 13 | 5500 | ESI+ | 3.51 |
| 3 | isorhamnetin-3-O-beta-d-glucoside | 479.3 | 317 | 253 | 150 | 34 | 10 | 13 | 5500 | ESI+ | 3.53 |
| 4 | tangeretin | 372.8 | 343.1 | 358.1 | 10 | 32 | 10 | 13 | 5500 | ESI+ | 4.67 |
| 5 | rutin | 609 | 299.9 | 279.5 | −50 | −43 | −10 | −15 | −4500 | ESI− | 3.44 |
| 6 | quercetin | 301 | 150.8 | 178.9 | −100 | −35 | −10 | −15 | −4500 | ESI− | 3.69 |
| 7 | isorhamnetin | 314.8 | 300 | 150.8 | −150 | −30 | −10 | −15 | −4500 | ESI− | 3.84 |
| 8 | quercetin-7-O-glucoside | 462.9 | 300.9 | 342.9 | −50 | −28 | −10 | −15 | −4500 | ESI− | 3.43 |
| 9 | kaempferol-3-O-glucoside | 592.9 | 284.9 | 255 | −50 | −40 | −10 | −15 | −4500 | ESI− | 3.50 |
| No. | Name | Linearity | LOD (ng/mL) | LOQ (ng/mL) | Stability (RSD, %) | Precision (RSD, %) | Repeatability (RSD, %) | ||
|---|---|---|---|---|---|---|---|---|---|
| Regression Equations | R2 | Ranges (ng/mL) | |||||||
| 1 | nobiletin | y = 62,293,800x − 1,576,600 | 0.9968 | 7.81–1000 | 0.488 | 0.977 | 3.05 | 1.43 | 4.44 |
| 2 | narcissin | y = 3,029,650x − 314,384 | 0.9942 | 31.25–4000 | 7.81 | 15.63 | 3.26 | 3.22 | 2.26 |
| 3 | isorhamnetin-3-O- beta-d-glucoside | y = 279,387x − 11,500 | 0.9973 | 62.5–4000 | 15.63 | 31.25 | 4.22 | 4.37 | 4.93 |
| 4 | tangeretin | y = 4,034,570x − 18,086 | 0.9902 | 3.91–250 | 0.488 | 0.977 | 1.94 | 1.76 | 4.87 |
| 5 | rutin | y = 3,089,740x − 61,316 | 0.9978 | 15.63–1000 | 7.81 | 31.25 | 2.99 | 3.71 | 1.74 |
| 6 | quercetin | y = 17,556,400x − 169,184 | 0.9921 | 15.63–500 | 7.81 | 15.63 | 3.82 | 4.03 | 4.9 |
| 7 | isorhamnetin | y = 116,434,300x − 149,754 | 0.9903 | 7.81–250 | 3.91 | 7.81 | 2.53 | 2.94 | 2.82 |
| 8 | quercetin-7-O- glucoside | y = 5,857,970x − 144,514 | 0.9989 | 31.25–1000 | 7.81 | 15.63 | 3.26 | 3.31 | 2.39 |
| 9 | kaempferol-3-O- glucoside | y = 4,499,260x − 117,172 | 0.9901 | 31.25–1000 | 7.81 | 15.63 | 2.06 | 4.49 | 4.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Y.; Chen, X.; Meng, Z.; Guo, S. Combined Metabolome and Transcriptome Analyses Reveal the Effects of Mycorrhizal Fungus Ceratobasidium sp. AR2 on the Flavonoid Accumulation in Anoectochilus roxburghii during Different Growth Stages. Int. J. Mol. Sci. 2020, 21, 564. https://doi.org/10.3390/ijms21020564
Zhang Y, Li Y, Chen X, Meng Z, Guo S. Combined Metabolome and Transcriptome Analyses Reveal the Effects of Mycorrhizal Fungus Ceratobasidium sp. AR2 on the Flavonoid Accumulation in Anoectochilus roxburghii during Different Growth Stages. International Journal of Molecular Sciences. 2020; 21(2):564. https://doi.org/10.3390/ijms21020564
Chicago/Turabian StyleZhang, Ying, Yuanyuan Li, Xiaomei Chen, Zhixia Meng, and Shunxing Guo. 2020. "Combined Metabolome and Transcriptome Analyses Reveal the Effects of Mycorrhizal Fungus Ceratobasidium sp. AR2 on the Flavonoid Accumulation in Anoectochilus roxburghii during Different Growth Stages" International Journal of Molecular Sciences 21, no. 2: 564. https://doi.org/10.3390/ijms21020564





