Transcriptome Modifications in the Porcine Intramuscular Adipocytes during Differentiation and Exogenous Stimulation with TNF-α and Serotonin
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Signatures of PIP Cells Differentiation
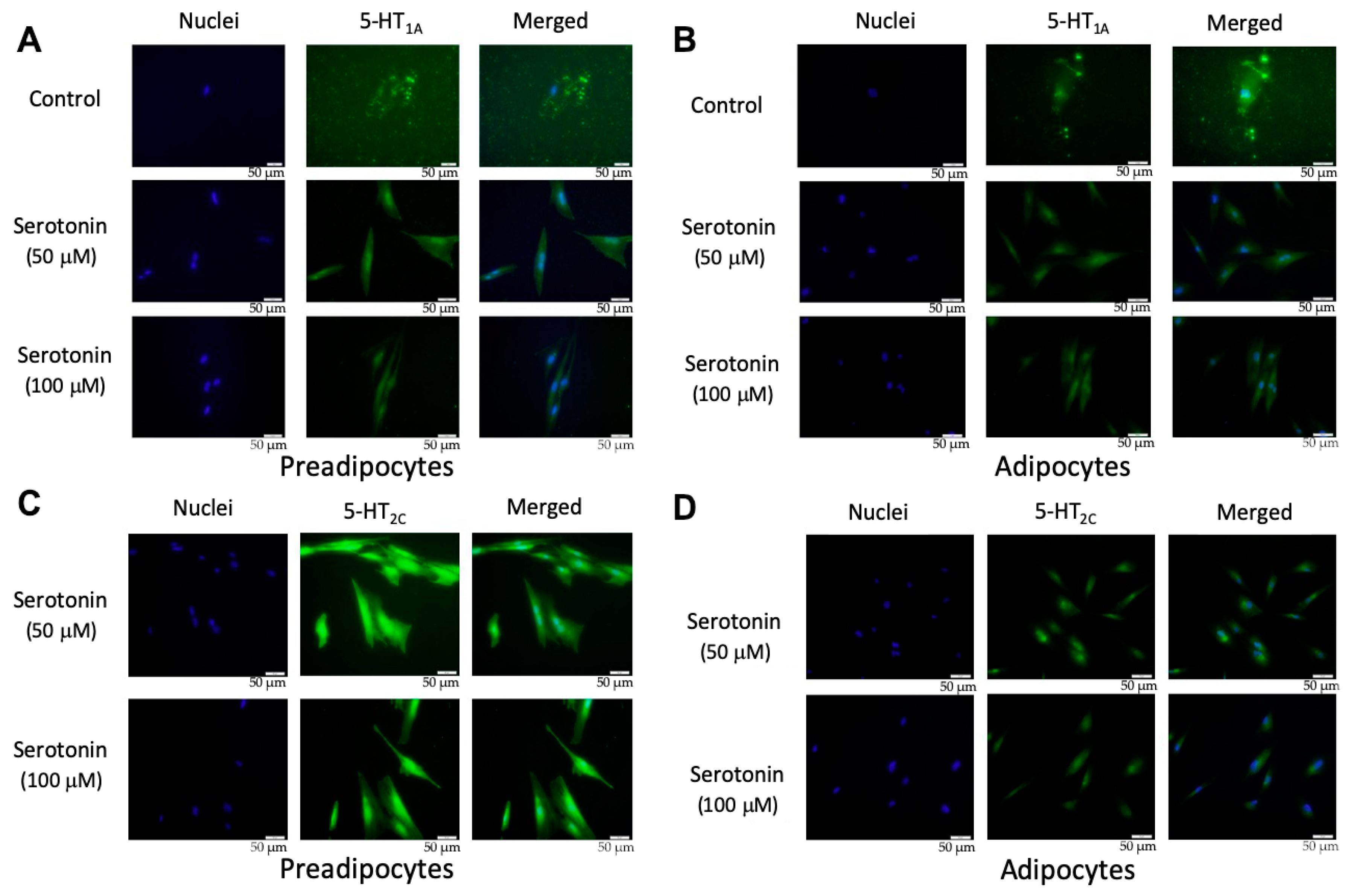
2.2. Expression of Serotonin Receptor Proteins in PIP and pMA
2.3. Transcriptome Modifications in pMA after Serotonin- or TNF-α -Stimulation
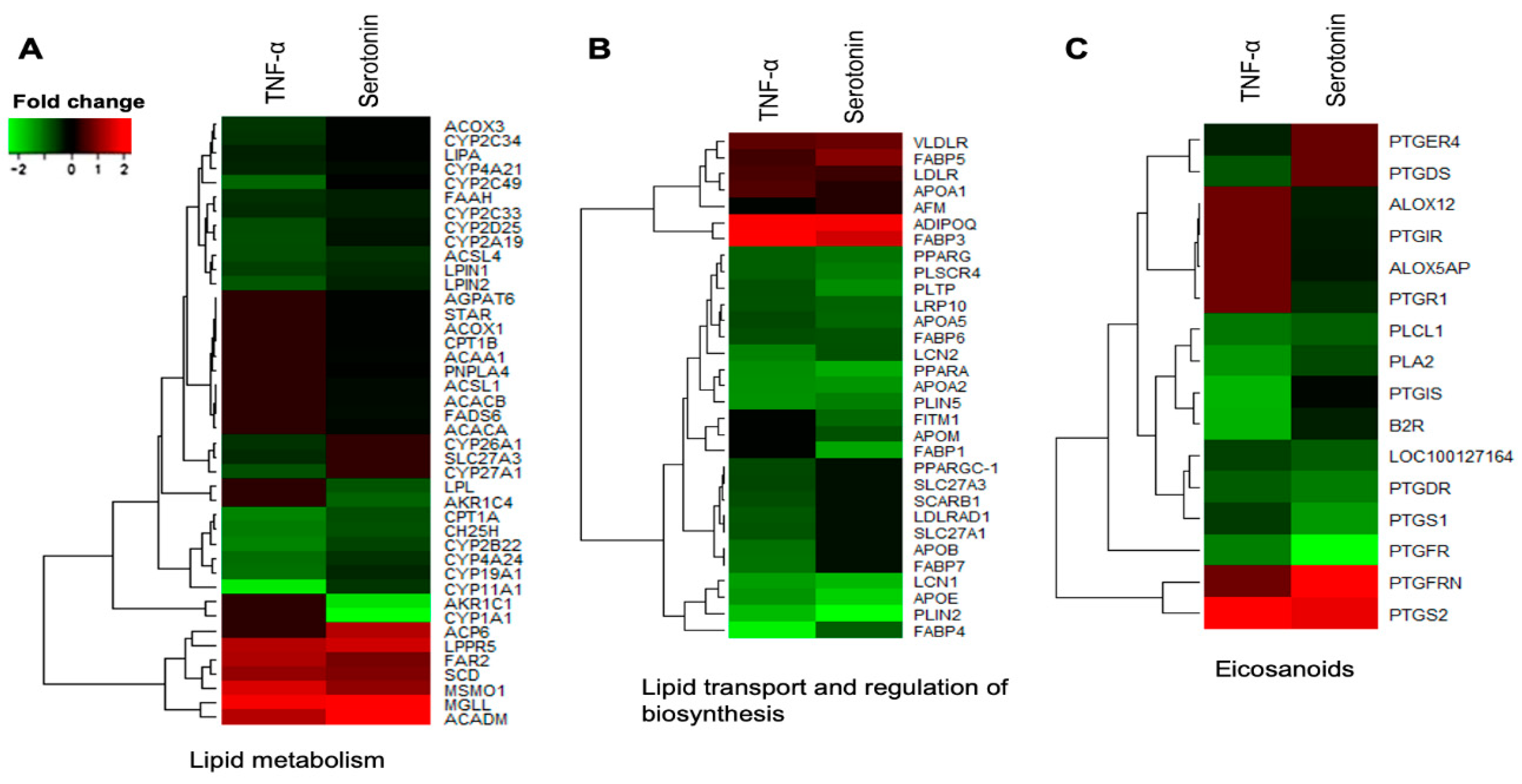
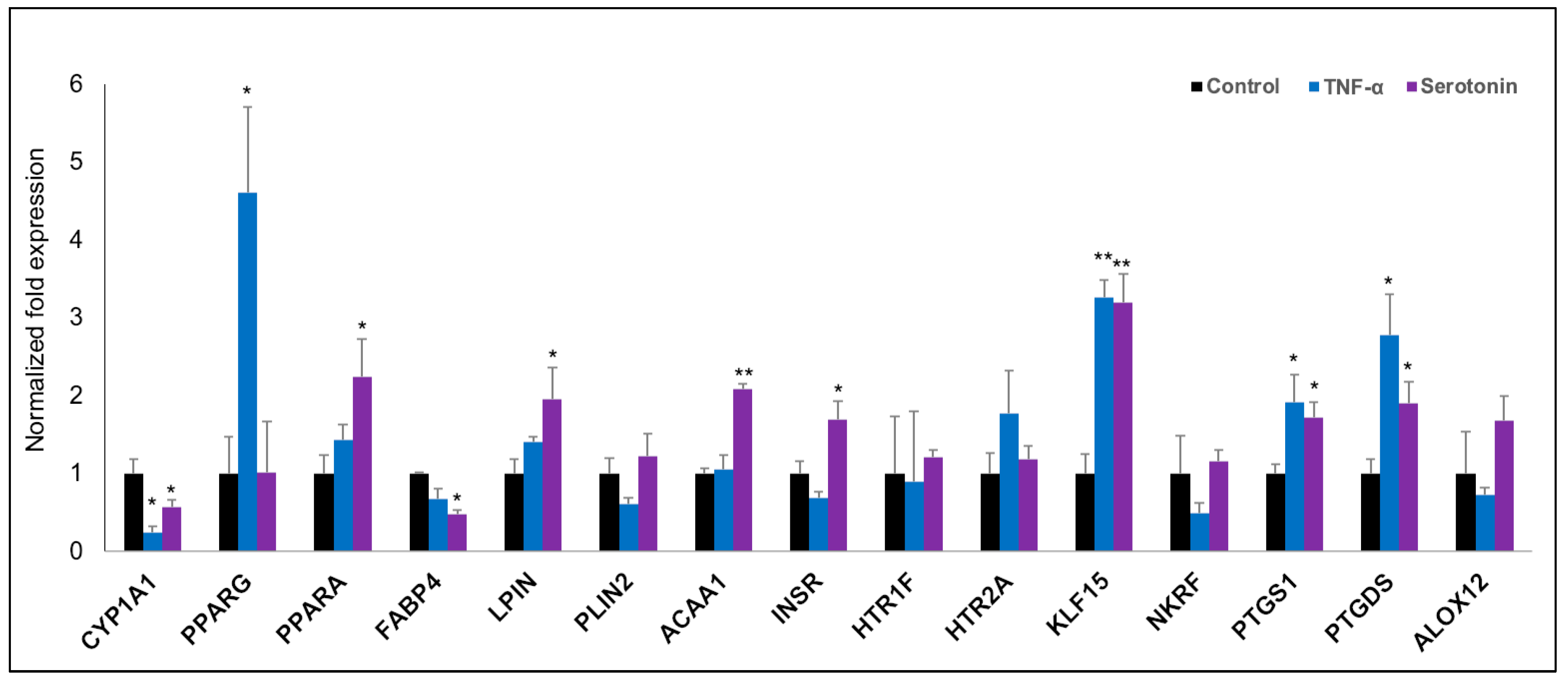
2.3.1. Differential Expressions of Metabolic and Endocrine Genes in pMA after Serotonin- or TNF-α -Stimulation
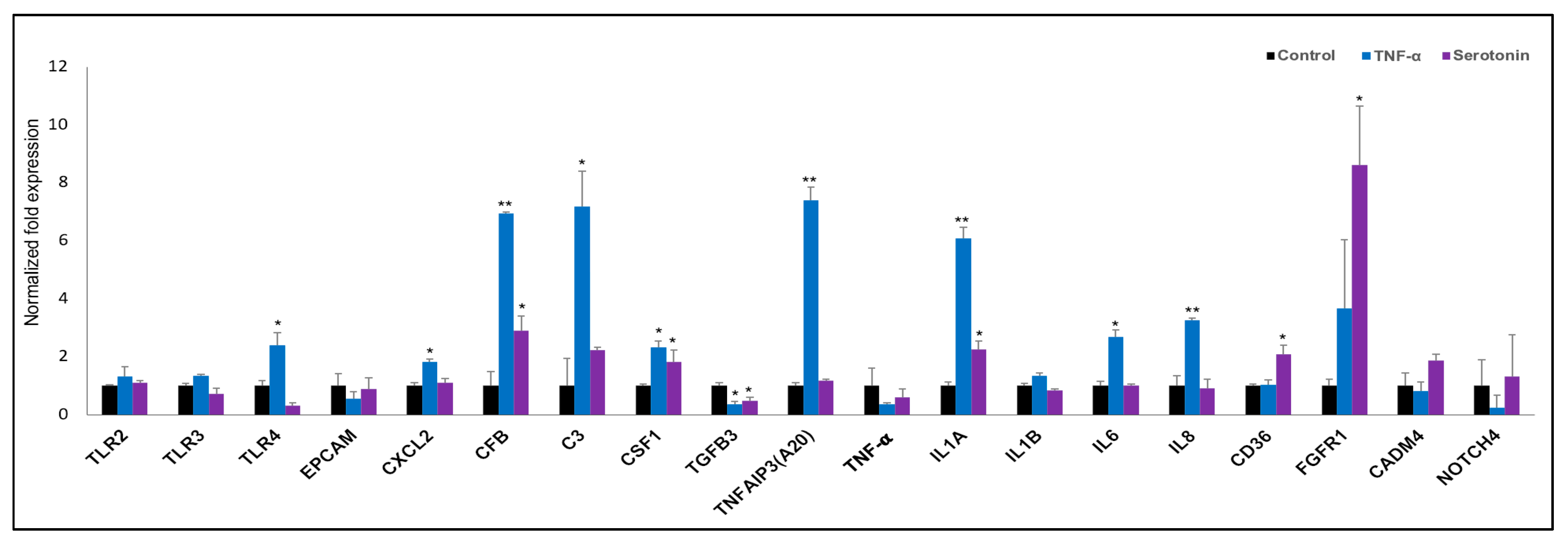
2.3.2. Differential Expressions of Immune Genes in pMA after Serotonin- or TNF-α -Stimulation
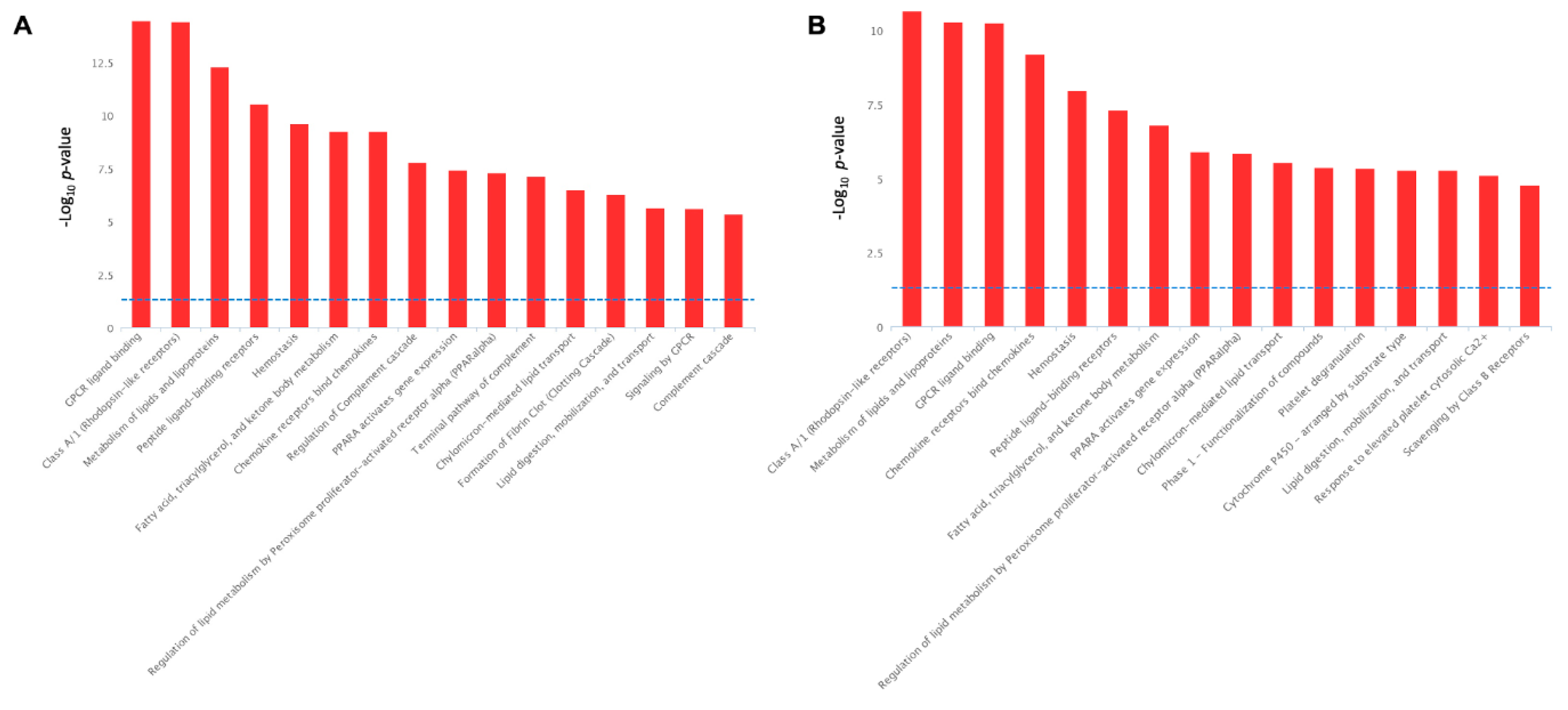
2.4. Pathways Activation by Serotonin- and TNF-α-Induced Transcriptome Alterations
3. Discussion
4. Materials and Methods
4.1. Cell Line and Culture Conditions
4.2. Differentiation of PIP into Mature Adipocytes
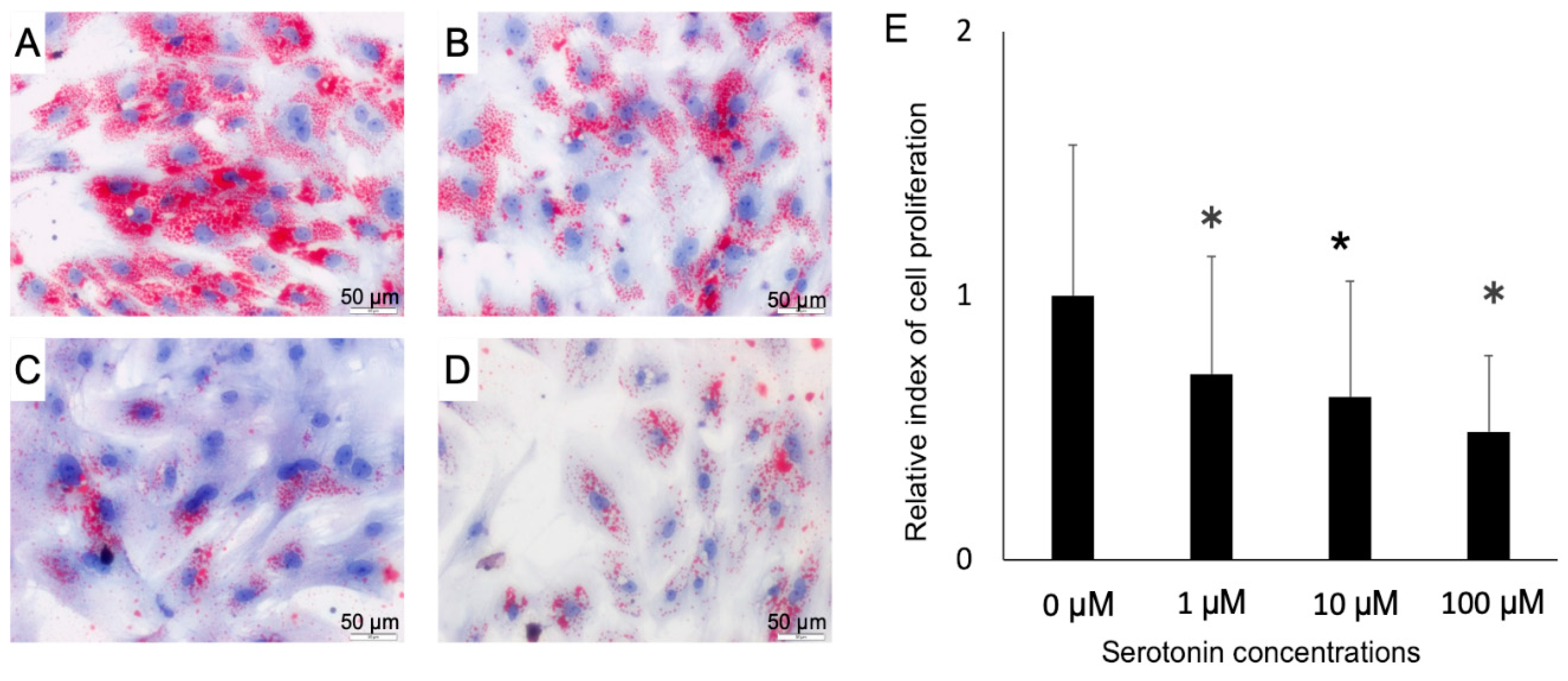
4.3. Proliferation Assay and Oil Red O Staining of Adipocytes
4.4. Immunofluorescent Staining
4.5. Stimulation of Adipocytes with Serotonin and TNF-α
4.6. RNA Isolation and Quality Control
4.7. RNA Labeling and Microarray Hybridization
4.8. Statistical Analysis of Microarray Data
4.9. Pathway Analysis
4.10. Network Analysis
4.11. Validation of Microarray Expression by RT-qPCR
4.12. Statistical Analysis of RT-qPCR Data
5. Conclusions
Supplementary Materials
Data Availability
Author Contributions
Funding
Conflicts of Interest
References
- Fischer-Posovszky, P.; Wabitsch, M.; Hochberg, Z. Endocrinology of adipose tissue—An update. Horm. Metab. Res. 2007, 39, 314–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, K.; Kusminski, C.M.; Scherer, P.E. Adipose tissue remodeling and obesity. J. Clin. Investig. 2011, 121, 2094–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samocha-Bonet, D.; Dixit, V.D.; Kahn, C.R.; Leibel, R.L.; Lin, X.; Nieuwdorp, M.; Pietilainen, K.H.; Rabasa-Lhoret, R.; Roden, M.; Scherer, P.E.; et al. Metabolicallyhealthy and unhealthy obese—The 2013 Stock Conference report. Obes. Rev. 2014, 15, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Villalon, C.M.; Centurion, D. Cardiovascular responses produced by 5- hydroxytriptamine: A pharmacological update on the receptors/mechanisms involved and therapeutic implications. Naunyn Schmiedebergs Arch. Pharmacol. 2007, 376, 45–63. [Google Scholar] [CrossRef]
- Gershon, M.D. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Mauler, M.; Bode, C.; Duerschmied, D. Platelet serotonin modulates immune functions. Hamostaseologie 2015, 35, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Crane, J.D.; Palanivel, R.; Mottillo, E.P.; Bujak, A.L.; Wang, H.; Ford, R.J.; Collins, A.; Blümer, R.M.; Fullerton, M.D.; Yabut, J.M.; et al. Inhibiting peripheral serotonin synthesis reduces obesity and metabolic dysfunction by promoting brown adipose tissue thermogenesis. Nat. Med. 2015, 21, 166–172. [Google Scholar] [CrossRef] [Green Version]
- Oh, C.M.; Namkung, J.; Go, Y.; Shong, K.E.; Kim, K.; Kim, H.; Park, B.Y.; Lee, H.W.; Jeon, Y.H.; Song, J.; et al. Regulation of systemic energy homeostasis by serotonin in adipose tissues. Nat. Commun. 2015, 6, 6794. [Google Scholar] [CrossRef]
- Hansson, L.; Stjernswärd, S.; Svensson, B. Changes in attitudes, intended behaviour, and mental health literacy in the Swedish population 2009–2014: An evaluation of a national antistigma programme. Acta Psychiatr. Scand. 2016, 134, 71–79. [Google Scholar] [CrossRef]
- Stunes, A.K.; Reseland, J.E.; Hauso, O.; Kidd, M.; Tommeras, K.; Waldum, H.L.; Syversen, U.; Gustafsson, B.I. Adipocytes express a functional system for serotonin synthesis, reuptake and receptor activation. Diabetes Obes. Metab. 2011, 13, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Welford, R.; Vercauteren, M.; Trébaul, A.; Cattaneo, C.; Eckert, D.; Garzotti, M.; Sieber, P.; Segrestaa, J.; Studer, R.; Groenen, P.M.; et al. Serotonin biosynthesis as a predictive marker of serotonin pharmacodynamics and disease-induced dysregulation. Sci. Rep. 2016, 6, 30059. [Google Scholar] [CrossRef] [PubMed]
- Sumara, G.; Sumara, O.; Kim, J.K.; Karsenty, G. Gut-derived serotonin is a multifunctional determinant to fasting adaptation. Cell Metab. 2012, 16, 588–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, Y.; Tomaru, U.; Miyoshi, A.; Ito, T.; Fukaya, S.; Miyoshi, H.; Atsumi, T.; Ishizu, A. Overexpression of TNF-α converting enzyme promotes adipose tissue inflammation and fibrosis induced by high fat diet. Exp. Mol. Pathol. 2014, 97, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Hube, B.; Lee, H. Expression pattern of tumour necrosis factor receptors in subcutaneous and omental human adipose tissue: Role of obesity and non-insulin-dependent diabetes mellitus. Eur. J. Clin. Investig. 1999, 29, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Marco, L.; Chacón, M.R.; Maymó-Masip, E.; Barroso, E.; Salvadó, L.; Wabitsch, M.; Garrido-Sánchez, L.; Tinahones, F.J.; Palomer, X.; Vendrell, J.; et al. TNF-α inhibits PPARβ/δ activity and SIRT1 expression through NF-κB in human adipocytes. Biochimica et Biophysica Acta (BBA) Mol. Cell Biol. Lipids 2012, 1821, 1177–1185. [Google Scholar] [CrossRef]
- Ruan, H.; Hacohen, N.; Golub, T.R.; Van, P.L.; Lodish, H.F. Tumor Necrosis Factor-α Suppresses Adipocyte-Specific Genes and Activates Expression of Preadipocyte Genes in 3T3-L1 Adipocytes: Nuclear Factor-κB Activation by TNF-α Is Obligatory. Diabetes 2002, 51, 1319–1336. [Google Scholar] [CrossRef] [Green Version]
- Sanosaka, M.; Minashima, T.; Suzuki, K.; Watanabe, K.; Ohwada, S.; Hagio, A.; Rose, M.T.; Yamaguchi, T.; Aso, H. A combination of octanoate and oleate promotes in vitro differentiation of porcine intramuscular adipocytes. Comp. Biochem. Physiol. A 2008, 149, 285–292. [Google Scholar] [CrossRef]
- Igata, M.; Islam, M.A.; Tada, A.; Takagi, M.; Kober, A.K.M.H.; Albarracin, L.; Aso, H.; Ikeda-Ohtsubo, W.; Miyazawa, K.; Yoda, K.; et al. Transcriptome Modifications in Porcine Adipocytes via Toll-Like Receptors Activation. Front. Immunol. 2019, 10, 1180. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, S.; Wang, Z.; Zhu, X.; Shu, G.; Liao, W.; Yu, K.; Gao, P.; Xi, Q.; Wang, X.; et al. Global comparison of gene expression profiles between intramuscular and subcutaneous adipocytes of neonatal landrace pig using microarray. Meat Sci. 2010, 86, 440–450. [Google Scholar] [CrossRef]
- Jiang, S.; Wei, H.; Song, T.; Yang, Y.; Peng, J.; Jiang, S. Transcriptome comparison between porcine subcutaneous and intramuscular stromal vascular cells during adipogenic differentiation. PLoS ONE 2013, 8, e77094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, D.; Yu, K.; Chen, H.; Chen, H.; Chen, L.; Liu, X.; He, Z.; Cong, P.; Chen, Y. Transcriptome Landscape of Porcine Intramuscular Adipocytes during Differentiation. J. Agric. Food Chem. 2017, 65, 6317–6328. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, S.; Tan, Z.; Wang, Y.; Zhang, F.; Yang, T.; Liu, Y.; Ao, H.; Xing, K.; Wang, C. Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation. Genes 2019, 10, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, D.E.; Nichols, C.D. Serotonin receptors. Chem. Rev. 2008, 108, 1614–1641. [Google Scholar] [CrossRef] [PubMed]
- El-Merahbi, R.; Löffler, M.; Mayer, A.; Sumara, G. The roles of peripheral serotonin in metabolic homeostasis. FEBS Lett. 2015, 589, 1728–1734. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, M.; Ono, K.; Horie, T.; Nagao, K.; Nishi, H.; Kuwabara, Y.; Takanabe-Mori, R.; Hasegawa, K.; Kita, T.; Kimura, T. Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol. Endocrinol. 2010, 24, 1978–1987. [Google Scholar] [CrossRef] [Green Version]
- Wyler, S.C.; Lord, C.C.; Lee, S.; Elmquist, J.K.; Liu, C. Serotonergic Control of Metabolic Homeostasis. Front. Cell. Neurosci. 2017, 11, 277. [Google Scholar] [CrossRef] [Green Version]
- Yabut, J.M.; Crane, J.D.; Green, A.E.; Keating, D.J.; Khan, W.I.; Steinberg, G.R. Emerging Roles for Serotonin in Regulating Metabolism: New Implications for an Ancient Molecule. Endocr. Rev. 2019, 40, 1092–1107. [Google Scholar] [CrossRef]
- Berglund, E.D.; Liu, C.; Sohn, J.W.; Liu, T.; Kim, M.H.; Lee, C.E.; Vianna, C.R.; Williams, K.W.; Xu, Y.; Elmquist, J.K. Serotonin 2C receptors in pro-opiomelanocortin neurons regulate energy and glucose homeostasis. J. Clin. Investig. 2013, 123, 5061. [Google Scholar] [CrossRef] [Green Version]
- Peckett, A.J.; Wright, D.C.; Riddell, M.C. The effects of glucocorticoids on adipose tissue lipid metabolism. Metabolism 2011, 60, 1500–1510. [Google Scholar] [CrossRef]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowherd, R.M.; Lyle, R.E.; McGehee, R.E. Molecular regulation of adipocyte differentiation. Semin. Cell Dev. Biol. 1999, 10, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.L.; Xiong, S.F.; Sun, J.M.; Yang, S.L.; Gu, Z.Y.; Liu, T.S. Pedostratigraphy and paleomagnetism of a ~7.0 Ma eolian loess–red clay sequence at Lingtai, Loess Plateau, north-central China and the implications for paleomonsoon evolution. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1999, 152, 49–66. [Google Scholar] [CrossRef]
- McNeel, R.L.; Ding, S.T.; Smith, E.O.; Mersmann, H.J. Expression of porcine adipocyte transcripts during differentiation in vitro and in vivo. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000, 126, 291–302. [Google Scholar] [CrossRef]
- Schioth, H.B.; Fredriksson, R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 2005, 142, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Harmar, A.J.; Hills, R.A.; Rosser, E.M.; Jones, M.; Buneman, O.P.; Dunbar, D.R.; Greenhill, S.D.; Hale, V.A.; Sharman, J.L.; Bonner, T.I.; et al. IUPHAR-DB: The IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009, 37, D680–D685. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Iken, K.; Chheng, S.; Fargin, A.; Goulet, A.-C.; Kouassi, E. Serotonin upregulates mitogen-stimulated B lymphocyte proliferation through 5-HT1A receptors. Cell. Immunol. 1995, 163, 1–9. [Google Scholar] [CrossRef]
- Kubera, M.; Maes, M.; Kenis, G.; Kim, Y.; Lason, W. Effects of serotonin and serotonergic agonists and antagonists on the production of tumor necrosis factor alpha and interleukin-6. Psychiatry Res. 2005, 134, 251–258. [Google Scholar] [CrossRef]
- Duerschmied, D. Platelet serotonin promotes the recruitment of neutrophils to sites of acute inflammation in mice. Blood 2013, 121, 1008–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghia, J.E. Serotonin has a key role in pathogenesis of experimental colitis. Gastroenterology 2009, 137, 1649–1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cawthorn, W.P.; Sethi, J.K. TNF-alpha and adipocyte biology. FEBS Lett. 2008, 582, 117–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafson, B.; Gogg, S.; Hedjazifar, S.; Jenndahl, L.; Hammarstedt, A.; Smith, U. Inflammation and impaired adipogenesis in hypertrophic obesity in man. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E999–E1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karastergiou, K.; Mohamed-Ali, V. The autocrine and paracrine roles of adipokines. Mol. Cell. Endocrinol. 2010, 318, 69–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkan, M.C.; Hevener, A.L.; Greten, F.R.; Maeda, S.; Li, Z.W.; Long, J.M.; Wynshaw-Boris, A.; Poli, G.; Olefsky, J.; Karin, M. IKK-beta links inflammation to obesity-induced insulin resistance. Nat. Med. 2005, 11, 191–198. [Google Scholar] [CrossRef]
- Vallerie, S.N.; Hotamisligil, G.S. The role of JNK proteins in metabolism. Sci. Transl. Med. 2010, 2, 60rv5. [Google Scholar] [CrossRef]
- Tan, X.; Cao, Z.; Li, M.; Xu, E.; Wang, J.; Xiao, Y. TNF- downregulates CIDEC via MEK/ERK pathway in human adipocytes. Obesity 2016, 24, 1070–1080. [Google Scholar] [CrossRef] [Green Version]
- Hotamisligil, G.S.; Budavari, A.; Murray, D.; Spiegelman, B.M. Reduced tyrosine kinase activity of the insulin receptor in obesity-diabetes: Central role of tumor necrosis factor-alpha. J. Clin. Investig. 1994, 94, 1543–1549. [Google Scholar] [CrossRef]
- Laurencikiene, J.; van Harmelen, V.; Nordström, E.; Dicker, A.; Blomqvist, L.; Näslund, E.; Langin, D.; Arner, A.; Rydén, M. NF-kappaB is important for TNF-alpha-induced lipolysis in human adipocytes. J. Lipid Res. 2007, 48, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Hulver, M.W.; Berggren, J.R.; Cortright, R.N.; Dudek, R.W.; Thompson, R.P.; Pories, W.J.; MacDonald, K.G.; Cline, G.W.; Shulman, G.I.; Dohm, G.L.; et al. Skeletal muscle lipid metabolism with obesity. Am. J. Physiol. Endocrinol. Metab. 2002, 284, E741–E747. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using r and Bioconductor; Gentleman, R., Carey, V., Dudoit, S., Irizarry, R., Huber, W., Eds.; Springer: New York, NY, USA, 2005; pp. 397–442. [Google Scholar]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. InnateDB: Systems biology of innate immunity and beyond—Recent updates and continuing curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Benner, M.J.; Hancock, R.E. NetworkAnalyst--integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygard, A.-B.; Jorgensen, C.B.; Cirera, S.; Fredholm, M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 2007, 8, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baudouin, L.C. Static Analysis. In Proceedings of the First International Static Analysis Symposium, SAS’94, Namur, Belgium, 28–30 September 1994; Lecture Notes in Computer Science 864. Springer: Berlin/Heidelberg, Germany, 1994. ISBN 3-540-58485-4. [Google Scholar]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tada, A.; Islam, M.A.; Kober, A.H.; Fukuyama, K.; Takagi, M.; Igata, M.; Albarracin, L.; Ikeda-Ohtsubo, W.; Miyazawa, K.; Yoda, K.; et al. Transcriptome Modifications in the Porcine Intramuscular Adipocytes during Differentiation and Exogenous Stimulation with TNF-α and Serotonin. Int. J. Mol. Sci. 2020, 21, 638. https://doi.org/10.3390/ijms21020638
Tada A, Islam MA, Kober AH, Fukuyama K, Takagi M, Igata M, Albarracin L, Ikeda-Ohtsubo W, Miyazawa K, Yoda K, et al. Transcriptome Modifications in the Porcine Intramuscular Adipocytes during Differentiation and Exogenous Stimulation with TNF-α and Serotonin. International Journal of Molecular Sciences. 2020; 21(2):638. https://doi.org/10.3390/ijms21020638
Chicago/Turabian StyleTada, Asuka, Md Aminul Islam, AKM Humayun Kober, Kohtaro Fukuyama, Michihiro Takagi, Manami Igata, Leonardo Albarracin, Wakako Ikeda-Ohtsubo, Kenji Miyazawa, Kazutoyo Yoda, and et al. 2020. "Transcriptome Modifications in the Porcine Intramuscular Adipocytes during Differentiation and Exogenous Stimulation with TNF-α and Serotonin" International Journal of Molecular Sciences 21, no. 2: 638. https://doi.org/10.3390/ijms21020638