Proteome Analysis of Walnut Bacterial Blight Disease
Abstract
:1. Introduction
2. Results
2.1. Quantitative Proteomic Analysis of Juglans regia in Response to Walnut Blight
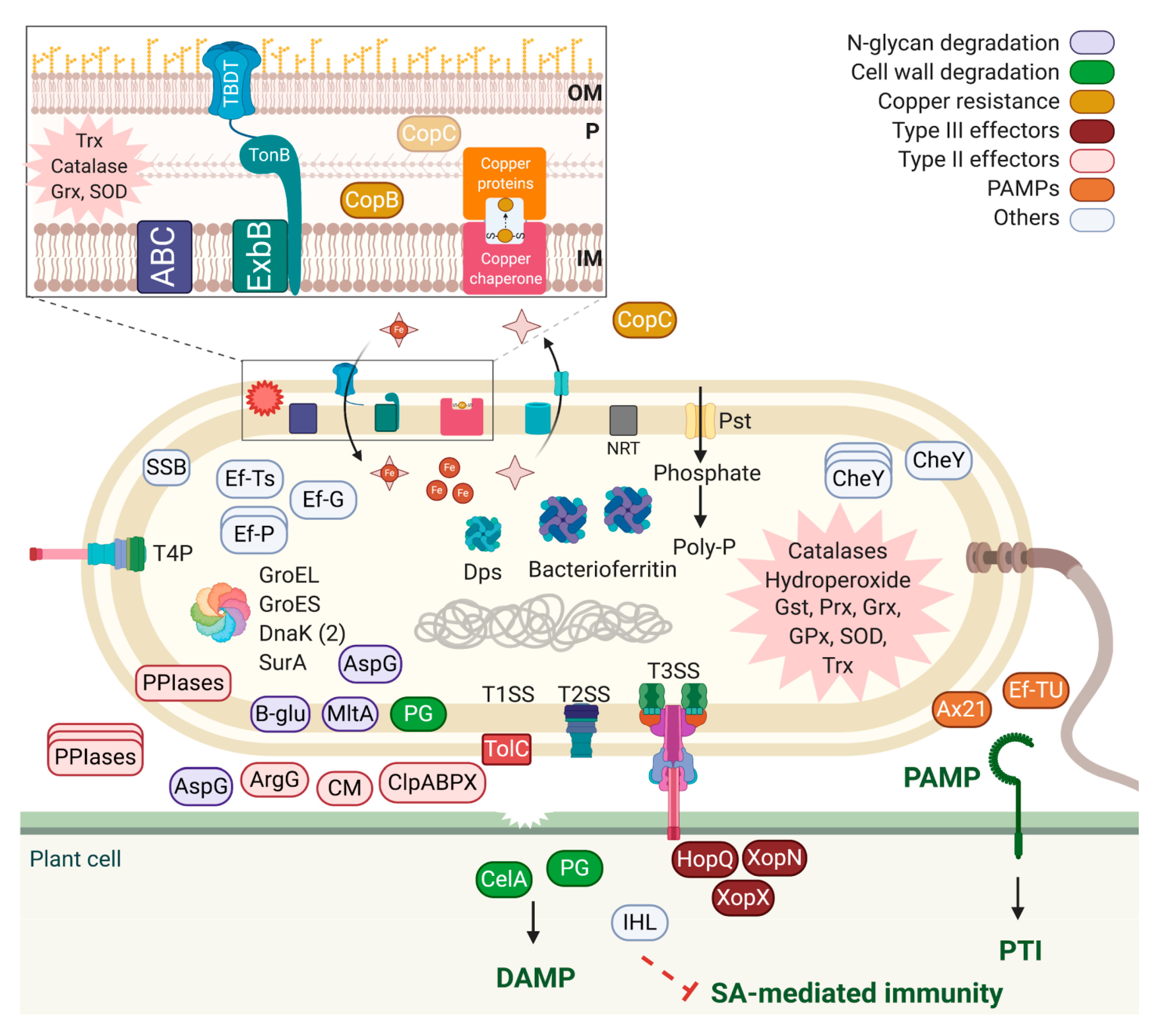
2.2. Quantitative Proteomic Analysis of Xanthomonas arboricola pv. juglandis in Walnut Hulls
2.3. Gene Ontology Annotation and Enrichment Analysis
2.4. Subcellular Localization
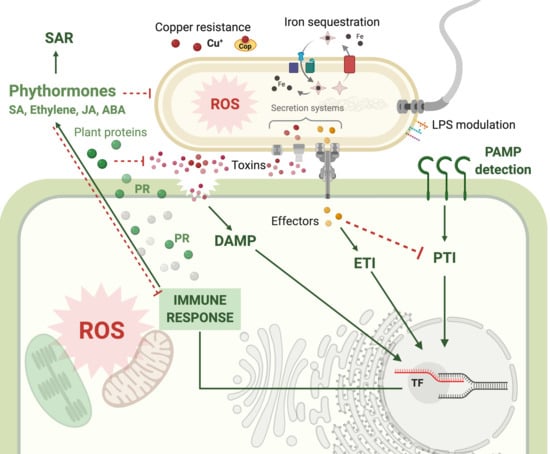
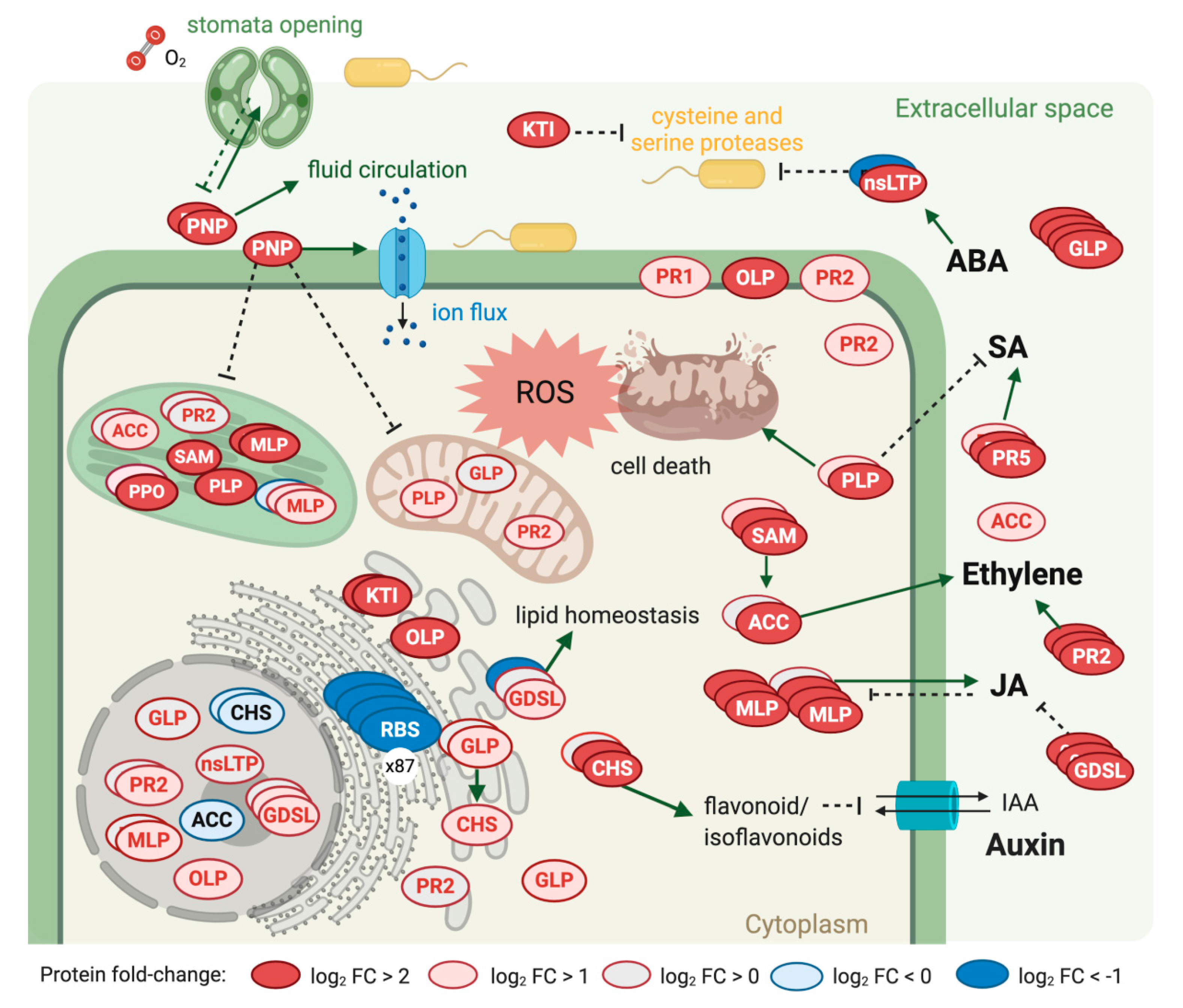
2.5. Plant Defense Response to Walnut Bacterial Blight Disease
3. Discussion
4. Materials and Methods
4.1. Plant Material and Nut Inoculations with Xanthomonas Arboricola pv. Juglandis 417
4.2. Scanning Electron Microscopy
4.3. Protein Extraction and Quantification
4.4. Protein Digestion, Tandem Mass Tag Labeling, and Mass-Spectrometry
4.5. Proteomic Analysis Workflow Pipeline and Statistical Analysis
4.6. Protein Functional Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Taha, N.A. Utility and importance of walnut, Juglans regia Linn: A review. Afr. J. Microbiol. Res. 2011, 5, 5796–5805. [Google Scholar]
- Pollegioni, P.; Woeste, K.; Chiocchini, F.; Del Lungo, S.; Ciolfi, M.; Olimpieri, I.; Tortolano, V.; Clark, J.; Hemery, G.; Mapelli, S.; et al. Rethinking the history of common walnut (Juglans regia L.) in Europe: Its origins and human interactions. PLoS ONE 2017, 12, e0172541. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Barreneche, T.; Lheureux, F.; Dirlewanger, E. Analysis of genetic diversity and structure in a worldwide walnut (Juglans regia L.) germplasm using SSR markers. PLoS ONE 2018, 13, e0208021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuts & Dried Fruits Statistical Yearbook 2017/2018; International Nut & Dried Fruit Council: Reus, Tarragona, Spain, 2017.
- Martínez, M.L.; O Labuckas, D.; Lamarque, A.L.; Maestri, D.M. Walnut (Juglans regia L.): Genetic resources, chemistry, by-products. J. Sci. Food Agric. 2010, 90, 1959–1967. [Google Scholar] [CrossRef]
- California Walnuts. Available online: https://walnuts.org/ (accessed on 2 September 2020).
- Robbins, W.A.; Xun, L.; Fitzgerald, L.Z.; Esguerra, S.; Henning, S.M.; Carpenter, C.L. Walnuts improve semen quality in men consuming a western-style diet: Randomized control dietary intervention trial. Biol. Reprod. 2012, 87, 101. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.-X.; Manchester, L.C.; Korkmaz, A.; Fuentes-Broto, L.; Hardman, W.E.; Rosales-Corral, S.A.; Qi, W. A walnut-enriched diet reduces the growth of LNCaP human prostate cancer xenografts in nude mice. Cancer Investig. 2013, 31, 365–373. [Google Scholar] [CrossRef] [Green Version]
- Kris-Etherton, P.M. Walnuts decrease risk of cardiovascular disease: A summary of efficacy and biologic mechanisms. J. Nutr. 2014, 144, 547S–554S. [Google Scholar] [CrossRef] [Green Version]
- Poulose, S.M.; Miller, M.G.; Shukitt-Hale, B. Role of walnuts in maintaining brain health with age. J. Nutr. 2014, 144, 561S–566S. [Google Scholar] [CrossRef] [Green Version]
- Hardman, W.E. Walnuts have potential for cancer prevention and treatment in mice. J. Nutr. 2014, 144, 555S–5560S. [Google Scholar] [CrossRef] [Green Version]
- Hagan, A.K.; Chiuve, E.S.; Stampfer, M.J.; Katz, J.N.; Grodstein, F. Greater adherence to the alternative healthy eating index is associated with lower incidence of physical function impairment in the nurses’ health study. J. Nutr. 2016, 146, 1341–1347. [Google Scholar] [CrossRef]
- Bitok, E.; Rajaram, S.; Jaceldo-Siegl, K.; Oda, K.; Sala-Vila, A.; Serra-Mir, M.; Ros, E.; Sabaté, J. Effects of long-term walnut supplementation on body weight in free-living elderly: Results of a randomized controlled trial. Nutrients 2018, 10, 1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holscher, H.D.; Guetterman, H.M.; Swanson, K.S.; An, R.; Matthan, N.R.; Lichtenstein, A.H.; Novotny, A.J.; Baer, D.J. Walnut consumption alters the gastrointestinal microbiota, microbially derived secondary bile acids, and health markers in healthy adults: A randomized controlled trial. J. Nutr. 2018, 148, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J.; et al. Primary prevention of cardiovascular disease with a Mediterranean diet supplemented with extra-virgin olive oil or nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef] [PubMed]
- Al Wattar, B.H.; Dodds, J.; Placzek, A.; Beresford, L.; Spyreli, E.; Moore, A.; Carreras, F.J.G.; Austin, F.; Murugesu, N.; Roseboom, T.J.; et al. Mediterranean-style diet in pregnant women with metabolic risk factors (ESTEEM): A pragmatic multicentre randomised trial. PLoS Med. 2019, 16, e1002857. [Google Scholar] [CrossRef]
- Lindow, S.; Olson, W.; Buchner, R. Colonization of dormant walnut buds by Xanthomonas arboricola pv. juglandis is predictive of subsequent disease. Phytopathology 2014, 104, 1163–1174. [Google Scholar] [CrossRef] [Green Version]
- Esterio, M.A.; Latorre, B.A. Potential sources of inoculum of Xanthomonas juglandis in Walnut Blight outbreaks. J. Hortic. Sci. 1982, 57, 69–72. [Google Scholar] [CrossRef]
- Frutos, D.; López, G. Search for Juglans regia genotypes resistant/tolerant to Xanthomonas arboricola pv. juglandis in the framework of cost action 873. J. Plant Pathol. 2012, 94, 1–37. [Google Scholar]
- Agricultural Telemetry Network & Consulting. Available online: http://www.agtelemetry.com/ (accessed on 3 September 2020).
- Olson, W.H.; Buchner, R.P. Leading edge of plant protection for walnuts. HortTechnology 2002, 12, 615–618. [Google Scholar] [CrossRef] [Green Version]
- Lamichhane, J.R.; Osdaghi, E.; Behlau, F.; Kohl, J.; Jones, J.B.; Aubertot, J.-N. Thirteen decades of antimicrobial copper compounds applied in agriculture. A review. Agron. Sustain. Dev. 2018, 38, 28. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-A. Increased toxicity of iron-amended copper-containing bactericides to the walnut blight pathogen Xanthomonas campestris pv. juglandis. Phytopathology 1993, 83, 1460. [Google Scholar] [CrossRef]
- Haack, S.E.; Wade, L.; Förster, H.; Adaskaveg, J.E. Epidemiology and management of bacterial spot of almond caused by Xanthomonas arboricola pv. pruni, a new disease in California. Plant Dis. 2020, 104, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schornack, S.; Meyer, A.; Römer, P.; Jordan, T.; Lahaye, T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 2006, 163, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Tamir-Ariel, D.; Navon, N.; Burdman, S. Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J. Bacteriol. 2007, 189, 6359–6371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, Y.-S.; Chern, M.; Bartley, L.E.; Han, M.; Jung, K.-H.; Lee, I.; Walia, H.; Richter, T.; Xu, X.; Cao, P.; et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011, 7, e1002020. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, A.; Zischek, C.; Lautier, M.; Jamet, S.; Rival, P.; Carrère, S.; Arlat, M.; Lauber, E. The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. mBio 2014, 5, e01527-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sels, J.; Mathys, J.; De Coninck, B.; Cammue, B.P.A.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Gottig, N.; Vranych, C.V.; Sgro, G.G.; Piazza, A.; Ottado, J. HrpE, the major component of the Xanthomonas type three protein secretion pilus, elicits plant immunity responses. Sci. Rep. 2018, 8, 9842. [Google Scholar] [CrossRef]
- Jiang, S.; Balan, B.; Assis, R.A.B.; Sagawa, C.H.D.; Wan, X.; Han, S.; Wang, L.; Zhang, L.; Zaini, P.A.; Walawage, S.L.; et al. Genome-wide profiling and phylogenetic analysis of the SWEET sugar transporter gene family in walnut and their lack of responsiveness to Xanthomonas arboricola pv. juglandis infection. Int. J. Mol. Sci. 2020, 21, 1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zekiri, F.; Molitor, C.; Mauracher, S.G.; Michael, C.; Mayer, R.L.; Gerner, C.; Rompel, A. Purification and characterization of tyrosinase from walnut leaves (Juglans regia). Phytochemistry 2014, 101, 5–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-García, P.J.; Crepeau, M.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016, 87, 507–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glagoleva, A.Y.; Shoeva, O.Y.; Khlestkina, E.K. Melanin pigment in plants: Current knowledge and future perspectives. Front. Plant Sci. 2020, 11, 770. [Google Scholar] [CrossRef]
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M.; et al. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014, 164, 1191–1203. [Google Scholar] [CrossRef] [Green Version]
- Pereira, U.P.; Gouran, H.; Nascimento, R.; Adaskaveg, J.E.; Goulart, L.R.; Dandekar, A.M. Complete genome sequence of Xanthomonas arboricola pv. juglandis 417, a copper-resistant strain isolated from Juglans regia L. Genome Announc. 2015, 3, e01126-15. [Google Scholar] [CrossRef] [Green Version]
- Marrano, A.; Britton, M.; Zaini, A.P.; Zimin, A.V.; Workman, E.R.; Puiu, D.; Bianco, L.; Di Pierro, E.A.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. GigaScience 2020, 9, giaa050. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucl. Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Li, L.; Xu, X.; Chen, C.; Shen, Z. Genome-wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis. Int. J. Mol. Sci. 2016, 17, 1622. [Google Scholar] [CrossRef] [Green Version]
- Sruthilaxmi, C.B.; Babu, S. Proteome responses to individual pathogens and abiotic conditions in rice seedlings. Phytopathology 2020, 110, 1326–1341. [Google Scholar] [CrossRef]
- Escobar, M.A.; Shilling, A.; Higgins, P.; Uratsu, S.L.; Dandekar, A.M. Characterization of polyphenol oxidase from walnut. J. Am. Soc. Hortic. Sci. 2008, 133, 852–858. [Google Scholar] [CrossRef] [Green Version]
- Li, S. Regulation of ribosomal proteins on viral infection. Cells 2019, 8, 508. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Donaldson, L.; Gehring, C.; Irving, H.R. Plant natriuretic peptides. Plant Signal. Behav. 2011, 6, 1606–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turek, I.S.; Marondedze, C.; Wheeler, J.I.; Gehring, C.; Irving, H.R. Plant natriuretic peptides induce proteins diagnostic for an adaptive response to stress. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottig, N.; Garavaglia, B.S.; Daurelio, L.; Valentine, A.J.; Gehring, C.; Orellano, E.G.; Ottado, J. Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide-like protein to modify host homeostasis. Proc. Natl. Acad. Sci. USA 2008, 105, 18631–18636. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a new osmotin-like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Houben, M.; Van De Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Wang, J.; Jia, H.; Kamran, A.; Qin, Y.; Liu, Y.; Hao, K.; Han, F.; Zhang, C.; Li, B.; et al. Identification and functional characterization of NbMLP28, a novel MLP-like protein 28 enhancing Potato virus Y resistance in Nicotiana benthamiana. BMC Microbiol. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell 1996, 8, 1809–1819. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Soliman, A.; Islam, R.; Adam, L.R.; Daayf, F. Verticillium dahliae’s isochorismatase hydrolase is a virulence factor that contributes to interference with potato’s salicylate and jasmonate defense signaling. Front. Plant Sci. 2017, 8, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Confer, A.W.; Ayalew, S. The OmpA family of proteins: Roles in bacterial pathogenesis and immunity. Vet. Microbiol. 2013, 163, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Dandekar, A.M.; Gouran, H.; Ibáñez, A.M.; Uratsu, S.L.; Agüero, C.B.; McFarland, S.; Borhani, Y.; Feldstein, P.A.; Bruening, G.; Nascimento, R.; et al. An engineered innate immune defense protects grapevines from Pierce disease. Proc. Natl. Acad. Sci. USA 2012, 109, 3721–3725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Kandel, P.P.; Cruz, L.F.; Cobine, P.A.; De La Fuente, L. the major outer membrane protein mopB is required for twitching movement and affects biofilm formation and virulence in two Xylella fastidiosa strains. Mol. Plant Microbe Interact. 2017, 30, 896–905. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-Y.; Wu, C.-H.; Lin, J.-W.; Weng, S.-F.; Tseng, Y.-H. Mutation of the gene encoding a major outer-membrane protein in Xanthomonas campestris pv. campestris causes pleiotropic effects, including loss of pathogenicity. Microbiology 2010, 156, 2842–2854. [Google Scholar] [CrossRef] [Green Version]
- Cesbron, S.; Briand, M.; Essakhi, S.; Gironde, S.; Boureau, T.; Manceau, C.; Saux, M.F.-L.; Jacques, M.-A. Comparative genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Jha, G.; Rajeshwari, R.; Sonti, R.V. Bacterial type two secretion system secreted proteins: Double-edged swords for plant pathogens. Mol. Plant Microbe Interact. 2005, 18, 891–898. [Google Scholar] [CrossRef] [Green Version]
- Cianciotto, N.P.; White, R.C. expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 2017, 85, e00014-17. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.M.; Moreira, L.M.; Ferro, J.A.; Soares, M.R.; De Laia, M.L.; Varani, A.M.; De Oliveira, J.C.; Ferro, M.I.T. Unravelling potential virulence factor candidates in Xanthomonas citri. subsp. citri by secretome analysis. Peer J. 2016, 4, e1734. [Google Scholar] [CrossRef] [Green Version]
- Assis, R.A.B.; Polloni, L.C.; Patané, J.S.L.; Thakur, S.; Felestrino, É.B.; Diaz-Caballero, J.; Digiampietri, L.A.; Goulart, L.R.; Almeida, N.F.; Nascimento, R.; et al. Identification and analysis of seven effector protein families with different adaptive and evolutionary histories in plant-associated members of the Xanthomonadaceae. Sci. Rep. 2017, 7, 16133. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gupta, R.; Song, W.; Huh, H.-H.; Lee, S.E.; Wu, J.; Agrawal, G.K.; Rakwal, R.; Kang, K.Y.; Park, S.-R.; et al. Label-free quantitative secretome analysis of Xanthomonas oryzae pv. oryzae highlights the involvement of a novel cysteine protease in its pathogenicity. J. Proteom. 2017, 169, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Rajeshwari, R.; Jha, G.; Sonti, R.V. Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant Microbe Interact. 2005, 18, 830–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, A.; Hirata, H.; Tsuyumu, S. HrpG regulates type II secretory proteins in Xanthomonas axonopodis pv. citri. J. Gen. Plant Pathol. 2008, 74, 138–150. [Google Scholar] [CrossRef]
- Metz, M.; Dahlbeck, D.; Morales, C.Q.; Al Sady, B.; Clark, E.T.; Staskawicz, B.J. The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 2005, 41, 801–814. [Google Scholar] [CrossRef]
- Deb, S.; Ghosh, P.; Patel, H.K.; Sonti, R.V. Interaction of the Xanthomonas effectors XopQ and XopX results in induction of rice immune responses. Plant J. 2020, 1–19. [Google Scholar] [CrossRef]
- Cheong, H.; Kim, C.-Y.; Jeon, J.-S.; Lee, B.-M.; Moon, J.S.; Hwang, I. Xanthomonas oryzae pv. oryzae type III effector XopN Targets OsVOZ2 and a putative thiamine synthase as a virulence factor in rice. PLoS ONE 2013, 8, e73346. [Google Scholar] [CrossRef]
- Kim, J.-G.; Li, X.; Roden, J.A.; Taylor, K.W.; Aakre, C.D.; Su, B.; LaLonde, S.; Kirik, A.; Chen, Y.; Baranage, G.; et al. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 2009, 21, 1305–1323. [Google Scholar] [CrossRef] [Green Version]
- Behlau, F.; Canteros, B.I.; Minsavage, G.V.; Jones, J.B.; Graham, J.H. Molecular Characterization of copper resistance genes from Xanthomonas citri subsp. citriand Xanthomonas alfalfae subsp. citrumelonis. Appl. Environ. Microbiol. 2011, 77, 4089–4096. [Google Scholar] [CrossRef] [Green Version]
- Lugo, A.J.; Elibox, W.; Jones, J.B.; Ramsubhag, A. Copper resistance in Xanthomonas campestris pv. campestris affecting crucifers in Trinidad. Eur. J. Plant Pathol. 2012, 136, 61–70. [Google Scholar] [CrossRef]
- Roach, R.; Mann, R.; Gambley, C.; Shivas, R.; Chapman, T.; Rodoni, B. Pathogenicity and copper tolerance in Australian Xanthomonas species associated with bacterial leaf spot. Crop. Prot. 2020, 127, 104923. [Google Scholar] [CrossRef]
- Palumaa, P. Copper chaperones. The concept of conformational control in the metabolism of copper. FEBS Lett. 2013, 587, 1902–1910. [Google Scholar] [CrossRef] [Green Version]
- Djamei, A.; Schipper, K.; Rabe, F.; Ghosh, A.; Vincon, V.; Kahnt, J.; Osorio, S.; Tohge, T.; Fernie, A.R.; Feussner, I.; et al. Metabolic priming by a secreted fungal effector. Nat. Cell Biol. 2011, 478, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Degrassi, G.; Devescovi, G.; Bigirimana, J.; Venturi, V. Xanthomonas oryzae pv. oryzae XKK.12 Contains an AroQγ chorismate mutase that is involved in rice virulence. Phytopathology 2010, 100, 262–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardales, E.Y.; Moon, S.J.; Park, S.R.; Shin, D.; Byun, M.O.; Noh, T.H. Inactivation of argG, encoding argininosuccinate synthetase from Xanthomonas oryzae pv. oryzae, is involved in bacterial growth and virulence in planta. Can. J. Plant Pathol. 2009, 31, 368–374. [Google Scholar] [CrossRef]
- Gouran, H.; Gillespie, H.; Nascimento, R.; Chakraborty, S.; Zaini, P.A.; Jacobson, A.; Phinney, B.S.; Dolan, D.; Durbin-Johnson, B.P.; Antonova, E.S.; et al. The secreted protease PrtA controls cell growth, biofilm formation and pathogenicity in Xylella fastidiosa. Sci. Rep. 2016, 6, 31098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, S.K.; Rajeshwari, R.; Sharma, Y.; Sonti, R.V. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. oryzae exhibits similarity to non-fimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 2002, 46, 637–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Chem. Biol. 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Faurobert, M.; Pelpoir, E.; Chaïb, J.; Valerie, M.; Catherine, D.; Michel, Z.; Hervé, T. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Plant Proteomics 2006, 355, 9–14. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Wilmarth, P.A.; Riviere, M.A.; David, L.L. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Inform. 2009, 2, 223–234. [Google Scholar] [CrossRef] [Green Version]
- McDonald, W.H.; Tabb, D.L.; Sadygov, R.G.; MacCoss, M.J.; Venable, J.; Graumann, J.; Johnson, J.R.; Cociorva, D.; Yates, J.R. MS1, MS2, and SQT—Three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 2004, 18, 2162–2168. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; Jahan, T.A.; Hoopmann, M.R. Comet: An open-source MS/MS sequence database search tool. Proteomics 2012, 13, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Elias, E.J.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Chem. Biol. 2007, 4, 207–214. [Google Scholar] [CrossRef]
- Plubell, D.L.; Wilmarth, P.A.; Zhao, Y.; Fenton, A.M.; Minnier, J.; Reddy, A.P.; Klimek, J.; Yang, X.; David, L.L.; Pamir, N. Extended Multiplexing of Tandem mass tags (TMT) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol. Cell. Proteom. 2017, 16, 873–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]






| GenBank ID | Locus Xaj417 | Description | SignalP | BUSCA Prediction | Rank Abundance |
|---|---|---|---|---|---|
| AKU49410.1 | AKJ12_RS06245 | OOP family OmpA-OmpF porin | OTHER | Cytoplasm | 1 |
| AKU49462.1 | AKJ12_RS14455 | Elongation factor Tu | OTHER | Cytoplasm | 2 |
| AKU49820.1 | AKJ12_RS08545 | Molecular chaperone GroEL | OTHER | Cytoplasm | 3 |
| AKU51588.1 | AKJ12_RS18640 | Ax21 family protein | OTHER | Cytoplasm | 4 |
| AKU50680.1 | AKJ12_RS13495 | Siderophore | SP(Sec/SPI) | SP-Extracellular space | 5 |
| AKU49415.1 | AKJ12_RS06280 | Malate dehydrogenase | OTHER | Cytoplasm | 6 |
| AKU51241.1 | AKJ12_RS16600 | Glucose dehydrogenase | OTHER | Cytoplasm | 7 |
| AKU48616.1 | AKJ12_RS01595 | 30S ribosomal protein S1 | OTHER | Cytoplasm | 8 |
| AKU51756.1 | AKJ12_RS19625 | Tetratricopeptide repeat protein | OTHER | Cytoplasm | 9 |
| AKU51362.1 | AKJ12_RS17320 | VirK-like protein | OTHER | Cytoplasm | 10 |
| AKU50394.1 | AKJ12_RS11890 | TonB-dependent receptor | SP(Sec/SPI) | SP-Extracellular space | 11 |
| AKU52070.1 | AKJ12_RS21505 | Hypothetical protein | OTHER | OM-Beta Strand | 12 |
| AKU51605.1 | AKJ12_RS18740 | Glutamine synthetase | OTHER | PM-Alpha Helix | 13 |
| AKU49821.1 | AKJ12_RS08550 | Molecular chaperone GroES | OTHER | Cytoplasm | 14 |
| AKU50456.1 | AKJ12_RS12245 | Peroxiredoxin | OTHER | PM-Alpha Helix | 15 |
| AKU51052.1 | AKJ12_RS15520 | ATP F0F1 synthase subunit alpha | OTHER | PM-Alpha Helix | 16 |
| AKU50723.1 | AKJ12_RS16695 | Succinyl-CoA synthetase subunit alpha | OTHER | Cytoplasm | 17 |
| AKU49376.1 | AKJ12_RS06020 | DNA-binding protein HU | OTHER | Cytoplasm | 18 |
| AKU49010.1 | AKJ12_RS03895 | Cold-shock protein | SP(Sec/SPI) | SP-Extracellular space | 19 |
| AKU50240.1 | AKJ12_RS10980 | Peptidylprolyl isomerase | OTHER | Cytoplasm | 20 |
| Protein Family | Code | Copies | Ave. Ratio log2 (Inf./Mock) | Direction | Ave. FDR |
|---|---|---|---|---|---|
| EG45-like domain-containing protein | PNP | 3 | 4.07 | up | 1.05 × 10−26 |
| Kunitz trypsin inhibitor | KTI | 3 | 4.05 | up | 2.30 × 10−15 |
| Pathogenesis-related 5 | PR5 | 3 | 3.46 | up | 9.96 × 10−22 |
| Osmotin-like protein | OLP | 3 | 2.89 | up | 9.72 × 10−19 |
| S-adenosylmethionine synthase | SAM | 4 | 2.04 | up | 2.71 × 10−13 |
| Major latex like-protein | MLP | 15 | 1.87 | up | 3.92 × 10−3 |
| Patatin-like protein | PLP | 4 | 1.84 | up | 1.08 × 10−4 |
| 1,3-beta-glucosidase | PR2 | 11 | 1.71 | up | 6.14 × 10−2 |
| Germin-like protein | GLP | 10 | 1.69 | up | 8.26 × 10−3 |
| GDSL esterase/lipase | GDSL | 14 | 1.14 | up | 1.23 × 10−1 |
| Pathogenesis-related 1 | PR1 | 1 | 1.14 | up | 3.67 × 10−7 |
| Chalcone synthase | CHS | 5 | 0.99 | up | 5.96 × 10−3 |
| 1-aminocyclopropane-1-carboxylate oxidase | ACC | 9 | 0.69 | up | 9.48 × 10−2 |
| Non-specific lipid-transfer protein | nsLPT | 6 | −0.46 | down | 2.09 × 10−2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
H. D. Sagawa, C.; de A. B. Assis, R.; Zaini, P.A.; Wilmarth, P.A.; Phinney, B.S.; Moreira, L.M.; Dandekar, A.M. Proteome Analysis of Walnut Bacterial Blight Disease. Int. J. Mol. Sci. 2020, 21, 7453. https://doi.org/10.3390/ijms21207453
H. D. Sagawa C, de A. B. Assis R, Zaini PA, Wilmarth PA, Phinney BS, Moreira LM, Dandekar AM. Proteome Analysis of Walnut Bacterial Blight Disease. International Journal of Molecular Sciences. 2020; 21(20):7453. https://doi.org/10.3390/ijms21207453
Chicago/Turabian StyleH. D. Sagawa, Cíntia, Renata de A. B. Assis, Paulo A. Zaini, Phillip A. Wilmarth, Brett S. Phinney, Leandro M. Moreira, and Abhaya M. Dandekar. 2020. "Proteome Analysis of Walnut Bacterial Blight Disease" International Journal of Molecular Sciences 21, no. 20: 7453. https://doi.org/10.3390/ijms21207453