Epigenetics and Heart Failure
Abstract
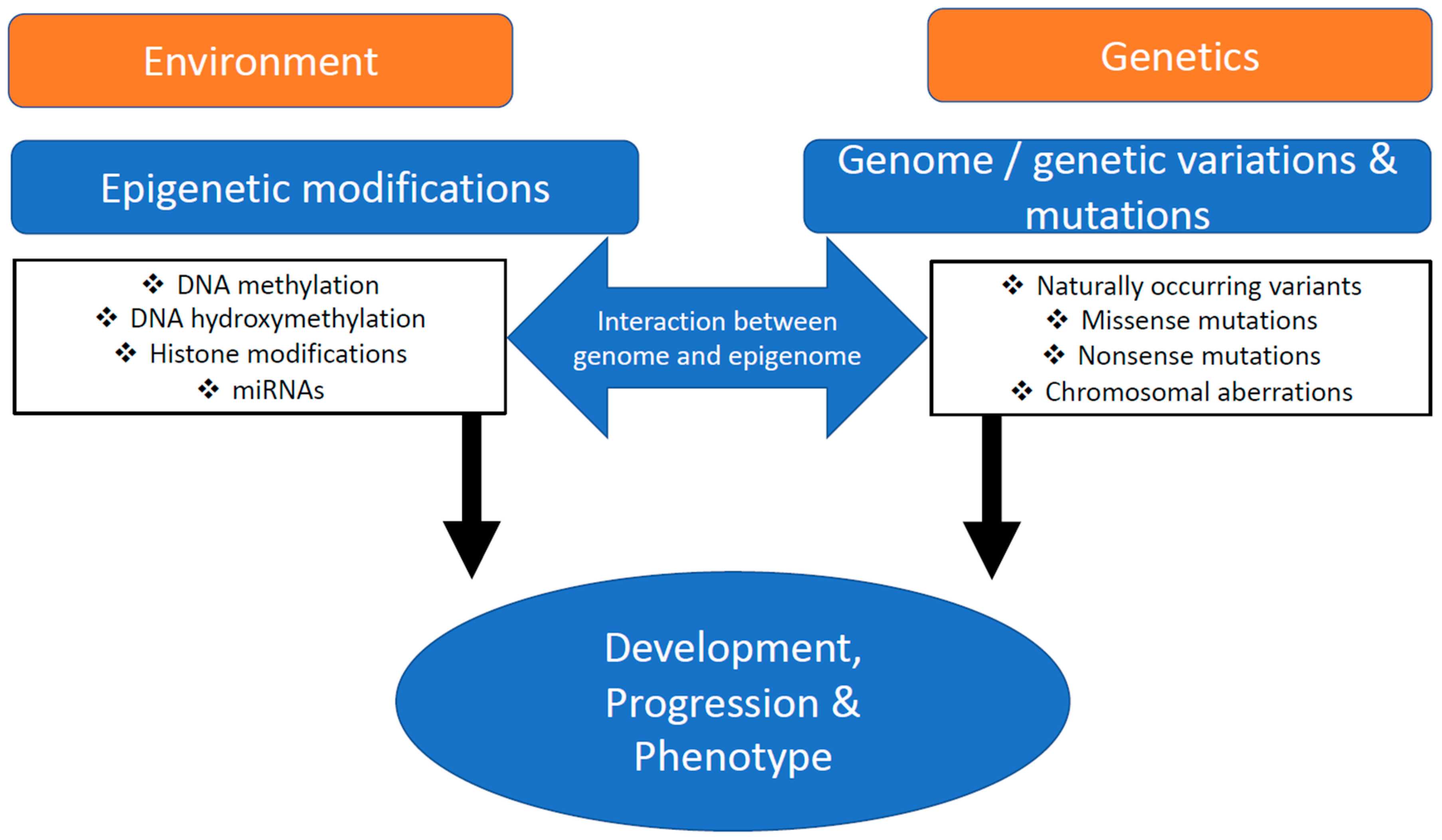
:1. Introduction
2. Transcription Factors and Chromatin Crosstalk in Experimental Heart Failure
3. Toward Epigenetic Hallmarks in Human Heart Failure
4. Conclusions and Outlook
Funding
Conflicts of Interest
Abbreviations
| TAC | Transverse aortic constriction |
| Csrp3 (Mlp) | Cysteine rich protein 3 |
| Mlp (Csrp3) | Muscle LIM protein |
| HATs | Histone acetyltransferases |
| HDACs | Deacetylases |
| GE | Gene expression |
References
- Lee, D.S.; Pencina, M.J.; Benjamin, E.J.; Wang, T.J.; Levy, D.; O’Donnell, C.J.; Nam, B.H.; Larson, M.G.; D’Agostino, R.B.; Vasan, R.S. Association of parental heart failure with risk of heart failure in offspring. N. Engl. J. Med. 2006, 355, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kathiresan, S.; Srivastava, D. Genetics of human cardiovascular disease. Cell 2012, 148, 1242–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marian, A.J.; Belmont, J. Strategic approaches to unraveling genetic causes of cardiovascular diseases. Circ. Res. 2011, 108, 1252–1269. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. The Epigenotpye; Endeavour: Sydney, Australia, 1942; pp. 18–20. [Google Scholar]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Ficz, G.; Branco, M.R.; Seisenberger, S.; Santos, F.; Krueger, F.; Hore, T.A.; Marques, C.J.; Andrews, S.; Reik, W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011, 473, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Pape, U.J.; Huang, Y.; Henderson, H.R.; Lister, R.; Ko, M.; McLoughlin, E.M.; Brudno, Y.; Mahapatra, S.; Kapranov, P.; et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011, 473, 394–397. [Google Scholar] [CrossRef] [Green Version]
- Gan, H.; Wen, L.; Liao, S.; Lin, X.; Ma, T.; Liu, J.; Song, C.X.; Wang, M.; He, C.; Han, C.; et al. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 2013, 4, 1995. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, M.; Kals, M.; Kacevska, M.; Barragan, I.; Kasuga, K.; Rane, A.; Metspalu, A.; Milani, L.; Ingelman-Sundberg, M. Ontogeny, distribution and potential roles of 5-hydroxymethylcytosine in human liver function. Genome Biol. 2013, 14, R83. [Google Scholar] [CrossRef] [Green Version]
- Tsagaratou, A.; Aijo, T.; Lio, C.W.; Yue, X.; Huang, Y.; Jacobsen, S.E.; Lahdesmaki, H.; Rao, A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E3306–E3315. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; D’Alessio, A.C.; Ito, S.; Wang, Z.; Cui, K.; Zhao, K.; Sun, Y.E.; Zhang, Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011, 25, 679–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.X.; Han, J.W.; Kim, S.; Namburi, S.; Hermetz, K.; Kim, J.J.; Rudd, M.K.; et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011, 7, e1002154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Flaherty, M.P.; Wu, W.J.; Tan, W.; Zhu, X.; Li, Q.; Bolli, R. Genetic background, gender, age, body temperature, and arterial blood pH have a major impact on myocardial infarct size in the mouse and need to be carefully measured and/or taken into account: Results of a comprehensive analysis of determinants of infarct size in 1,074 mice. Basic Res. Cardiol. 2012, 107, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Orozco, L.D.; Wang, J.; Rau, C.D.; Rubbi, L.; Ren, S.; Wang, Y.; Pellegrini, M.; Lusis, A.J.; Vondriska, T.M. DNA Methylation Indicates Susceptibility to Isoproterenol-Induced Cardiac Pathology and Is Associated With Chromatin States. Circ. Res. 2016, 118, 786–797. [Google Scholar] [CrossRef] [Green Version]
- Greco, C.M.; Kunderfranco, P.; Rubino, M.; Larcher, V.; Carullo, P.; Anselmo, A.; Kurz, K.; Carell, T.; Angius, A.; Latronico, M.V.; et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat. Commun. 2016, 7, 12418. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [Green Version]
- Taegtmeyer, H.; Sen, S.; Vela, D. Return to the fetal gene program: A suggested metabolic link to gene expression in the heart. Ann. N. Y. Acad. Sci. 2010, 1188, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Razeghi, P.; Young, M.E.; Alcorn, J.L.; Moravec, C.S.; Frazier, O.H.; Taegtmeyer, H. Metabolic gene expression in fetal and failing human heart. Circulation 2001, 104, 2923–2931. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.P.; Tan, W.L.W.; Anene-Nzelu, C.G.; Lee, C.J.M.; Li, P.Y.; Luu, T.D.A.; Chan, C.X.; Tiang, Z.; Ng, S.L.; Huang, X.; et al. Robust CTCF-Based Chromatin Architecture Underpins Epigenetic Changes in the Heart Failure Stress-Gene Response. Circulation 2019, 139, 1937–1956. [Google Scholar] [CrossRef]
- Rosa-Garrido, M.; Chapski, D.J.; Schmitt, A.D.; Kimball, T.H.; Karbassi, E.; Monte, E.; Balderas, E.; Pellegrini, M.; Shih, T.T.; Soehalim, E.; et al. High-Resolution Mapping of Chromatin Conformation in Cardiac Myocytes Reveals Structural Remodeling of the Epigenome in Heart Failure. Circulation 2017, 136, 1613–1625. [Google Scholar] [CrossRef]
- Braunwald, E.; Zipes, L. Heart Disease—A Textbook of Cardiovascular Medicine, 10th Revised ed.; Saunders: Philadelphia, PA, USA, 2015. [Google Scholar]
- McNally, E.M.; Golbus, J.R.; Puckelwartz, M.J. Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Investig. 2013, 123, 19–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arber, S.; Hunter, J.J.; Ross, J., Jr.; Hongo, M.; Sansig, G.; Borg, J.; Perriard, J.C.; Chien, K.R.; Caroni, P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 1997, 88, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Knöll, R.; Hoshijima, M.; Hoffman, H.M.; Person, V.; Lorenzen-Schmidt, I.; Bang, M.L.; Hayashi, T.; Shiga, N.; Yasukawa, H.; Schaper, W.; et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 2002, 111, 943–955. [Google Scholar] [CrossRef] [Green Version]
- Geier, C.; Perrot, A.; Ozcelik, C.; Binner, P.; Counsell, D.; Hoffmann, K.; Pilz, B.; Martiniak, Y.; Gehmlich, K.; van der Ven, P.F.; et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation 2003, 107, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Bassel-Duby, R.; Olson, E.N. Control of cardiac growth and function by calcineurin signaling. J. Biol. Chem. 2003, 278, 36981–36984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey, N.; Richardson, J.A.; Olson, E.N. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 14632–14637. [Google Scholar] [CrossRef] [Green Version]
- Heineke, J.; Ruetten, H.; Willenbockel, C.; Gross, S.C.; Naguib, M.; Schaefer, A.; Kempf, T.; Hilfiker-Kleiner, D.; Caroni, P.; Kraft, T.; et al. Attenuation of cardiac remodeling after myocardial infarction by muscle LIM protein-calcineurin signaling at the sarcomeric Z-disc. Proc. Natl. Acad. Sci. USA 2005, 102, 1655–1660. [Google Scholar] [CrossRef] [Green Version]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Morales, C.R.; Gillette, T.G.; Hill, J.A. Epigenetic regulation in heart failure. Curr. Opin. Cardiol. 2016, 31, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Gupta, M.P.; Samant, S.A.; Smith, S.H.; Shroff, S.G. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J. Biol. Chem. 2008, 283, 10135–10146. [Google Scholar] [CrossRef] [Green Version]
- Paudyal, A.; Dewan, S.; Ikie, C.; Whalley, B.J.; de Tombe, P.P.; Boateng, S.Y. Nuclear accumulation of myocyte muscle LIM protein is regulated by heme oxygenase 1 and correlates with cardiac function in the transition to failure. J. Physiol. 2016, 594, 3287–3305. [Google Scholar] [CrossRef] [PubMed]
- Buyandelger, B.; Ng, K.E.; Miocic, S.; Piotrowska, I.; Gunkel, S.; Ku, C.H.; Knöll, R. MLP (muscle LIM protein) as a stress sensor in the heart. Pflug. Arch. 2011, 462, 135–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, S.P.; Altschuld, R.A. Increased glycolytic metabolism in cardiac hypertrophy and congestive failure. Am. J. Physiol. 1970, 218, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Feinendegen, L.E.; Henrich, M.M.; Kuikka, J.T.; Thompson, K.H.; Vester, E.G.; Strauer, B. Myocardial lipid turnover in dilated cardiomyopathy: A dual in vivo tracer approach. J. Nucl. Cardiol. 1995, 2, 42–52. [Google Scholar] [CrossRef]
- Moalic, J.M.; Charlemagne, D.; Mansier, P.; Chevalier, B.; Swynghedauw, B. Cardiac hypertrophy and failure—A disease of adaptation. Modifications in membrane proteins provide a molecular basis for arrhythmogenicity. Circulation 1993, 87, IV21–IV26. [Google Scholar] [PubMed]
- Wittels, B.; Spann, J.F., Jr. Defective lipid metabolism in the failing heart. J. Clin. Investig. 1968, 47, 1787–1794. [Google Scholar] [CrossRef] [Green Version]
- Sack, M.N.; Rader, T.A.; Park, S.; Bastin, J.; McCune, S.A.; Kelly, D.P. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation 1996, 94, 2837–2842. [Google Scholar] [CrossRef]
- Reiser, P.J.; Portman, M.A.; Ning, X.H.; Schomisch Moravec, C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H1814–H1820. [Google Scholar] [CrossRef] [Green Version]
- Hunter, J.J.; Chien, K.R. Signaling pathways for cardiac hypertrophy and failure. N. Engl. J. Med. 1999, 341, 1276–1283. [Google Scholar] [CrossRef]
- Puente, B.N.; Kimura, W.; Muralidhar, S.A.; Moon, J.; Amatruda, J.F.; Phelps, K.L.; Grinsfelder, D.; Rothermel, B.A.; Chen, R.; Garcia, J.A.; et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 2014, 157, 565–579. [Google Scholar] [CrossRef] [Green Version]
- Gilsbach, R.; Preissl, S.; Gruning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Wurch, A.; Bonisch, U.; Gunther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014, 5, 5288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilsbach, R.; Schwaderer, M.; Preissl, S.; Gruning, B.A.; Kranzhofer, D.; Schneider, P.; Nuhrenberg, T.G.; Mulero-Navarro, S.; Weichenhan, D.; Braun, C.; et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun. 2018, 9, 391. [Google Scholar] [CrossRef] [Green Version]
- van Duijvenboden, K.; de Bakker, D.E.M.; Man, J.C.K.; Janssen, R.; Gunthel, M.; Hill, M.C.; Hooijkaas, I.B.; van der Made, I.; van der Kraak, P.H.; Vink, A.; et al. Conserved NPPB+ Border Zone Switches From MEF2- to AP-1-Driven Gene Program. Circulation 2019, 140, 864–879. [Google Scholar] [CrossRef]
- Movassagh, M.; Choy, M.K.; Goddard, M.; Bennett, M.R.; Down, T.A.; Foo, R.S. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE 2010, 5, e8564. [Google Scholar] [CrossRef] [PubMed]
- Krenz, M.; Robbins, J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J. Am. Coll. Cardiol. 2004, 44, 2390–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molkentin, J.D.; Lu, J.R.; Antos, C.L.; Markham, B.; Richardson, J.; Robbins, J.; Grant, S.R.; Olson, E.N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 1998, 93, 215–228. [Google Scholar] [CrossRef] [Green Version]
- van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [Green Version]
- van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J., Jr.; Olson, E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef] [Green Version]
- Jabbar, A.; Pingitore, A.; Pearce, S.H.; Zaman, A.; Iervasi, G.; Razvi, S. Thyroid hormones and cardiovascular disease. Nat. Rev. Cardiol. 2017, 14, 39–55. [Google Scholar] [CrossRef]
- Meder, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Frese, K.; Lai, A.; Nietsch, R.; Scheiner, C.; Mester, S.; Bordalo, D.M.; et al. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation 2017, 136, 1528–1544. [Google Scholar] [CrossRef]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, K.K.; Nicholls, S.J.; Ginsberg, H.D.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Cummings, J.L.; Sweeney, M.; et al. Effect of selective BET protein inhibitor apabetalone on cardiovascular outcomes in patients with acute coronary syndrome and diabetes: Rationale, design, and baseline characteristics of the BETonMACE trial. Am. Heart J. 2019, 217, 72–83. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ameer, S.S.; Hossain, M.B.; Knöll, R. Epigenetics and Heart Failure. Int. J. Mol. Sci. 2020, 21, 9010. https://doi.org/10.3390/ijms21239010
Ameer SS, Hossain MB, Knöll R. Epigenetics and Heart Failure. International Journal of Molecular Sciences. 2020; 21(23):9010. https://doi.org/10.3390/ijms21239010
Chicago/Turabian StyleAmeer, Syeda Shegufta, Mohammad Bakhtiar Hossain, and Ralph Knöll. 2020. "Epigenetics and Heart Failure" International Journal of Molecular Sciences 21, no. 23: 9010. https://doi.org/10.3390/ijms21239010




