Dynamic Changes in pStat3 Are Involved in Meiotic Spindle Assembly in Mouse Oocytes
Abstract
1. Introduction
2. Results
2.1. Changes in Relative Stat3 and pStat3 Expression from Oocyte Maturation to Pre-Implantation Stages
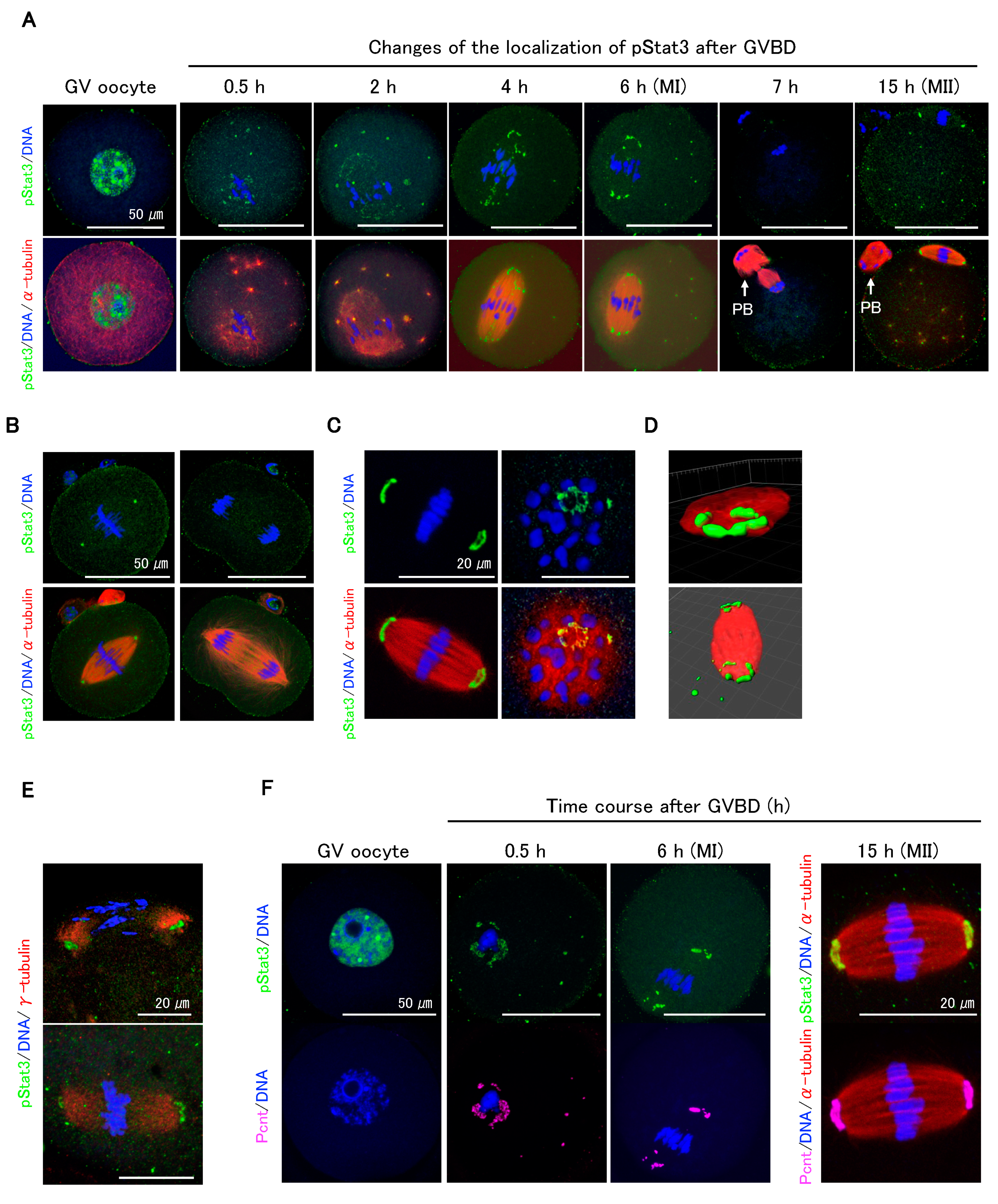
2.2. pStat3 Localization
2.3. Stat3/pStat3 Expression in Stat3−/− Oocytes
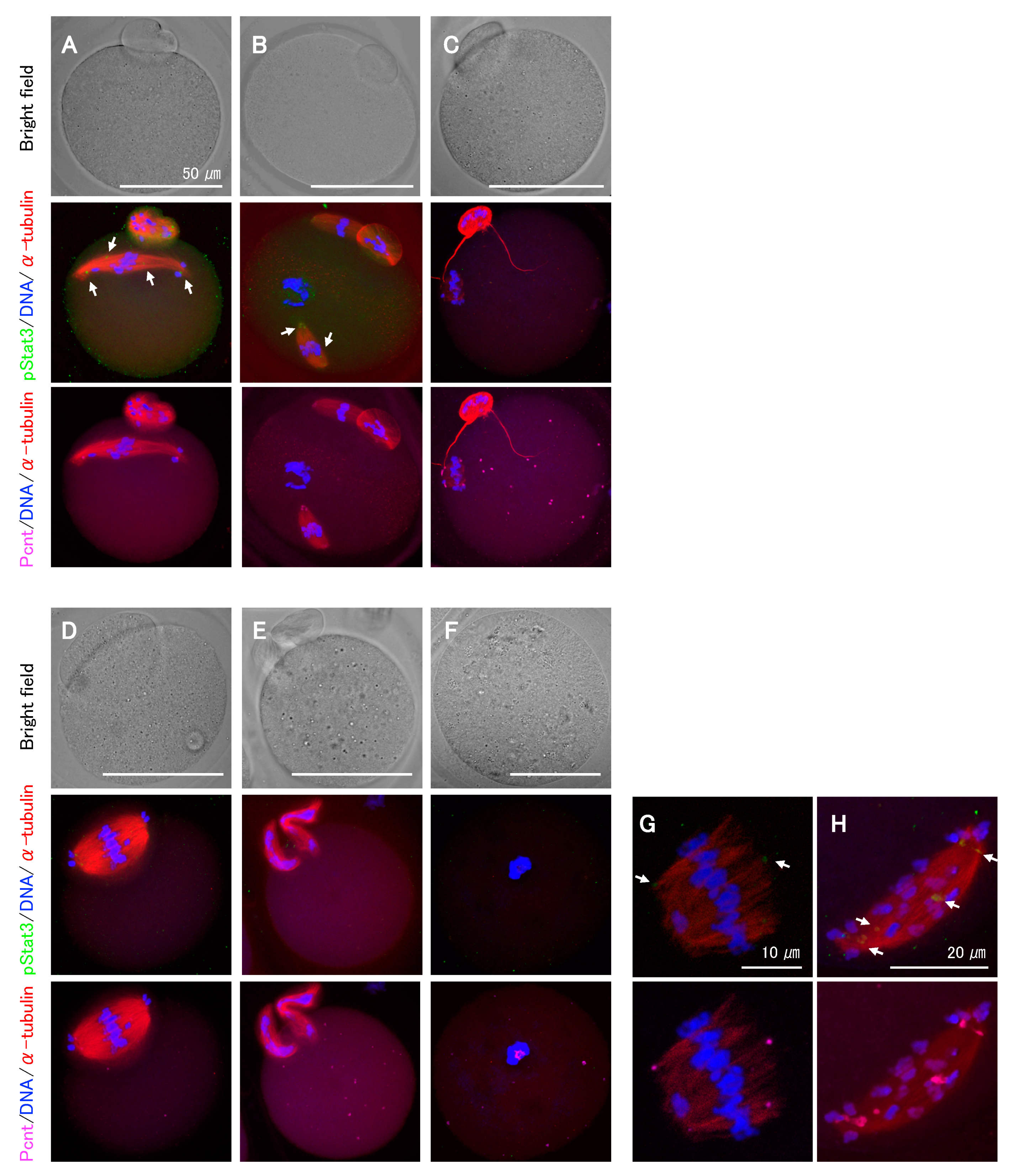
2.4. Effects of pStat3 Inhibition on Meiotic Spindle Assembly and Chromosome Segregation
2.5. Effects of pStat3 Disruption during IVM on Pre-Implantation Stage Embryos
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Genotyping
4.3. Collection of GV Oocytes and IVM
4.4. IVF
4.5. Evaluation of Small-Molecule Stat3 Inhibitors
4.6. Evaluation of Anti-pStat3 Antibody Injection
4.7. Western Blotting
4.8. Immunocytochemistry
4.9. RT-qPCR
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Ihle, J.N. Cytokine receptor signalling. Nature 1995, 377, 591–594. [Google Scholar] [CrossRef]
- Shuai, K.; Liu, B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003, 3, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Darnell, J.E., Jr. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Lütticken, C.; Wegenka, U.M.; Yuan, J.; Buschmann, J.; Schindler, C.; Ziemiecki, A.; Harpur, A.G.; Wilks, A.F.; Yasukawa, K.; Taga, T.; et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science 1994, 263, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Stahl, N.; Boulton, T.G.; Farruggella, T.; Ip, N.Y.; Davis, S.; Witthuhn, B.A.; Quelle, F.W.; Silvennoinen, O.; Barbieri, G.; Pellegrini, S.; et al. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science 1994, 263, 92–95. [Google Scholar] [CrossRef]
- Stahl, N.; Farruggella, T.J.; Boulton, T.G.; Zhong, Z.; Darnell, J.E., Jr.; Yancopoulos, G.D. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science 1995, 267, 1349–1353. [Google Scholar] [CrossRef]
- Ng, D.C.; Lin, B.H.; Lim, C.P.; Huang, G.; Zhang, T.; Poli, V.; Cao, X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 2006, 172, 245–257. [Google Scholar] [CrossRef]
- Verma, N.K.; Dourlat, J.; Davies, A.M.; Long, A.; Liu, W.Q.; Garbay, C.; Kelleher, D.; Volkov, Y. STAT3-stathmin interactions control microtubule dynamics in migrating T-cells. J. Biol. Chem. 2009, 284, 12349–12362. [Google Scholar] [CrossRef]
- Morris, E.J.; Kawamura, E.; Gillespie, J.A.; Balgi, A.; Kannan, N.; Muller, W.J.; Roberge, M.; Dedhar, S. Stat3 regulates centrosome clustering in cancer cells via Stathmin/PLK1. Nat. Commun. 2017, 8, 15289. [Google Scholar] [CrossRef]
- Antczak, M.; Van Blerkom, J. Oocyte influences on early development: The regulatory proteins leptin and STAT3 are polarized in mouse and human oocytes and differentially distributed within the cells of the preimplantation stage embryo. Mol. Hum. Reprod. 1997, 3, 1067–1086. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Carvajal, L.; Medico, L.; Pepling, M. Expression of Stat3 in germ cells of developing and adult mouse ovaries and testes. Gene Expr. Patterns 2005, 5, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; de Matos, D.G.; Fan, H.Y.; Shimada, M.; Palmer, S.; Richards, J.S. Interleukin-6: An autocrine regulator of the mouse cumulus cell-oocyte complex expansion process. Endocrinology 2009, 150, 3360–3368. [Google Scholar] [CrossRef] [PubMed]
- Dang-Nguyen, T.Q.; Haraguchi, S.; Kikuchi, K.; Somfai, T.; Bodó, S.; Nagai, T. Leukemia inhibitory factor promotes porcine oocyte maturation and is accompanied by activation of signal transducer and activator of transcription 3. Mol. Reprod. Dev. 2014, 81, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Brower, P.T.; Gizang, E.; Boreen, S.M.; Schultz, R.M. Biochemical studies of mammalian oogenesis: Synthesis and stability of various classes of RNA during growth of the mouse oocyte in vitro. Dev. Biol. 1981, 86, 373–383. [Google Scholar] [CrossRef]
- De Leon, V.; Johnson, A.; Bachvarova, R. Half-lives and relative amounts of stored and polysomal ribosomes and poly(A) + RNA in mouse oocytes. Dev. Biol. 1983, 98, 400–408. [Google Scholar] [CrossRef]
- Morgan, M.; Much, C.; DiGiacomo, M.; Azzi, C.; Ivanova, I.; Vitsios, D.M.; Pistolic, J.; Collier, P.; Moreira, P.N.; Benes, V.; et al. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 2017, 548, 347–351. [Google Scholar] [CrossRef]
- Bouniol-Baly, C.; Hamraoui, L.; Guibert, J.; Beaujean, N.; Szöllösi, M.S.; Debey, P. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol. Reprod. 1999, 60, 580–587. [Google Scholar] [CrossRef]
- De La Fuente, R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev. Biol. 2006, 292, 1–12. [Google Scholar] [CrossRef]
- Hodgman, R.; Tay, J.; Mendez, R.; Richter, J.D. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development 2001, 128, 2815–2822. [Google Scholar]
- De La Fuente, R.; Eppig, J.J. Transcriptional activity of the mouse oocyte genome: Companion granulosa cells modulate transcription and chromatin remodeling. Dev. Biol. 2001, 229, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef] [PubMed]
- de Vries, W.N.; Binns, L.T.; Fancher, K.S.; Dean, J.; Moore, R.; Kemler, R.; Knowles, B.B. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis 2000, 26, 110–112. [Google Scholar] [CrossRef]
- Lan, Z.J.; Xu, X.; Cooney, A.J. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 2004, 71, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Do, D.V.; Ueda, J.; Messerschmidt, D.M.; Lorthongpanich, C.; Zhou, Y.; Feng, B.; Guo, G.; Lin, P.J.; Hossain, M.Z.; Zhang, W.; et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013, 27, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Robker, R.L.; Watson, L.N.; Robertson, S.A.; Dunning, K.R.; McLaughlin, E.A.; Russell, D.L. Identification of sites of STAT3 action in the female reproductive tract through conditional gene deletion. PLoS ONE 2014, 9, e101182. [Google Scholar] [CrossRef]
- Moh, A.; Iwamoto, Y.; Chai, G.X.; Zhang, S.S.; Kano, A.; Yang, D.D.; Zhang, W.; Wang, J.; Jacoby, J.J.; Gao, B.; et al. Role of STAT3 in liver regeneration: Survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab. Invest. 2007, 87, 1018–1028. [Google Scholar] [CrossRef]
- Schust, J.; Sperl, B.; Hollis, A.; Mayer, T.U.; Berg, T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006, 13, 1235–1242. [Google Scholar] [CrossRef]
- Zhang, X.; Yue, P.; Page, B.D.; Li, T.; Zhao, W.; Namanja, A.T.; Paladino, D.; Zhao, J.; Chen, Y.; Gunning, P.T.; et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. USA 2012, 109, 9623–9628. [Google Scholar] [CrossRef]
- Vitale, S.G.; Rossetti, P.; Corrado, F.; Rapisarda, A.M.; La Vignera, S.; Condorelli, R.A.; Valenti, G.; Sapia, F.; Laganà, A.S.; Buscema, M. How to Achieve High-Quality Oocytes? The Key Role of Myo-Inositol and Melatonin. Int. J. Endocrinol. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Dumont, J.; Desai, A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012, 22, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Bennabi, I.; Terret, M.E.; Verlhac, M.H. Meiotic spindle assembly and chromosome segregation in oocytes. J. Cell Biol. 2016, 215, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Carabatsos, M.J.; Combelles, C.M.; Messinger, S.M.; Albertini, D.F. Sorting and reorganization of centrosomes during oocyte maturation in the mouse. Microsc. Res. Tech. 2000, 49, 435–444. [Google Scholar] [CrossRef]
- Gueth-Hallonet, C.; Antony, C.; Aghion, J.; Santa-Maria, A.; Lajoie-Mazenc, I.; Wright, M.; Maro, B. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J. Cell Sci. 1993, 105, 157–166. [Google Scholar]
- Zimmerman, W.C.; Sillibourne, J.; Rosa, J.; Doxsey, S.J. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 2004, 15, 3642–3657. [Google Scholar] [CrossRef]
- Delaval, B.; Doxsey, S.J. Pericentrin in cellular function and disease. J. Cell Biol. 2010, 188, 181–190. [Google Scholar] [CrossRef]
- Meng, X.Q.; Fan, H.Y.; Zhong, Z.S.; Zhang, G.; Li, Y.L.; Chen, D.Y.; Sun, Q.Y. Localization of gamma-tubulin in mouse eggs during meiotic maturation, fertilization, and early embryonic development. J. Reprod. Dev. 2004, 50, 97–105. [Google Scholar] [CrossRef][Green Version]
- Baumann, C.; Wang, X.; Yang, L.; Viveiros, M.M. Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J. Cell Sci. 2017, 130, 1251–1262. [Google Scholar] [CrossRef]
- Chen, C.T.; Hehnly, H.; Yu, Q.; Farkas, D.; Zheng, G.; Redick, S.D.; Hung, H.F.; Samtani, R.; Jurczyk, A.; Akbarian, S.; et al. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr. Biol. 2014, 24, 2327–2334. [Google Scholar] [CrossRef]
- Alonzi, T.; Maritano, D.; Gorgoni, B.; Rizzuto, G.; Libert, C.; Poli, V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol. Cell. Biol. 2001, 21, 1621–1632. [Google Scholar] [CrossRef]
- Toyoda, Y.; Yokoyama, M.; Hoshi, T. Studies on fertilisation of mouse eggs in vitro. I. In vitro fertilisation of eggs by fresh epididymal sperm. Jpn. J. Anim. Reprod. 1971, 16, 147–151. [Google Scholar]
- Kito, S.; Hayao, T.; Noguchi-Kawasaki, Y.; Ohta, Y.; Hideki, U.; Tateno, S. Improved in vitro fertilization and development by use of modified human tubal fluid and applicability of pronucleate embryos for cryopreservation by rapid freezing in inbred mice. Comp. Med. 2004, 54, 564–570. [Google Scholar] [PubMed]
- Takeo, T.; Nakagata, N. Reduced glutathione enhances fertility of frozen/thawed C57BL/6 mouse sperm after exposure to methyl-beta-cyclodextrin. Biol. Reprod. 2011, 85, 1066–1072. [Google Scholar] [CrossRef] [PubMed]
- Lawitts, J.A.; Biggers, J.D. Culture of preimplantation embryos. Methods Enzymol. 1993, 225, 153–164. [Google Scholar] [PubMed]


| Inhibitors | No. of Oocytes Examined | No. (%) of Oocytes Extruded 1st PB | No. (%) of Abnormal MII Oocyte* |
|---|---|---|---|
| (Stat3+/+) | |||
| Stattic | |||
| 0 μM** | 102 | 99 (97.2 ± 1.8)a | 3 (2.8 ± 1.8)a |
| 1 μM | 102 | 100 (98.0 ± 1.2)a | 10 (9.9 ± 2.8)a |
| 2 μM | 102 | 98 (96.2 ± 1.7)a | 45 (44.3 ± 6.3)b |
| 3 μM | 102 | 25 (24.8 ± 11.0)b | 93 (86.3 ± 4.3)c |
| 4 μM | 102 | 0 (0)c | 102 (100)c |
| BP-1-102 | |||
| 0 μM** | 100 | 99 (99.0 ± 1.0)a | 3 (3.0 ± 2.0)a |
| 2 μM | 100 | 100 (100)a | 5 (5 ± 1.6)a |
| 4 μM | 100 | 98 (96.2 ± 1.2)a | 13 (13 ± 3.4a |
| 8 μM | 100 | 96 (96.0 ± 1.9)a | 62 (62 ± 6.8)b |
| 16 μM | 100 | 82 (82.0 ± 5.1)b | 100 (100)c |
| (Stat−/−) | |||
| Stattic | |||
| 0 μM** | 64 | 62 (95.6 ± 4.4) a | 4 (6.5 ± 0.4) a |
| 1 μM | 59 | 58 (98.0 ± 2.0) a | 7 (12.5 ± 1.5) a |
| 2 μM | 62 | 56 (91.8 ± 2.0) a | 28 (45.8 ± 7.2) b |
| 3 μM | 60 | 13 (21.9 ±3.8) b | 53 (88.6 ± 4.0) c |
| 4 μM | 50 | 0 (0) c | 50 (100) d |
| Microinjection | No. of Oocytes Examined | No. (%) of Oocytes Extruded 1st PB | No. (%) of Abnormal MII Oocyte* |
|---|---|---|---|
| Anti-pStat3 antibody | 270 | 239 (87.8 ± 2.1)a | 72 (27.5 ± 2.5)a |
| Isotype control IgG | 245 | 237 (97.0 ± 1.2)ab | 9 (3.5 ±1.3)b |
| PBS | 213 | 208 (98.6 ± 1.1)b | 8 (4.4 ± 1.9)b |
| Inhibitors | No. of Oocytes Examined | No. (%) of Oocytes Extruded 1st PB | No. (%) of Embryos Developed to | |||
|---|---|---|---|---|---|---|
| 2-Cell | 4-Cell | 8-Cell/Morula | Blastocyst | |||
| Stattic | ||||||
| 0 μM * | 184 | 183 (99.4 ± 0.6) a | 164 (89.0 ± 3.3) a | 129 (69.9 ± 2.8) a | 123 (66.5 ± 3.8) a | 113 (61.2 ± 2.5) a |
| 1 μM | 188 | 186 (90.0 ± 0.7) a | 113 (60.0 ± 5.2) b | 35 (18.4 ± 1.9) b | 34 (17.9 ± 2.1) b | 31 (16.4 ± 2.8) b |
| 2 μM | 194 | 190 (97.9 ± 1.4) a | 114 (57.4 ± 5.3) b | 20 (9.7 ± 3.0) c | 18 (8.6 ± 2.7) c | 111 (5.2 ± 1.8) c |
| 3 μM | 199 | 70 (34.8 ± 9.8) b | 37 (18.4 ± 7.4) c | 0 (0) d | 0 (0) d | 0 (0) d |
| BP-1-102 | ||||||
| 0 μM * | 95 | 95 (100) | 83 (87.4 ± 4.7) a | 68 (71.9±3.3) a | 68 (71.9±3.3) a | 62 (65.3 ± 3.3) a |
| 2 μM | 105 | 104 (98.9 ± 1.1) | 88 (83.6 ± 8.2) ab | 52 (49.3±5.4) b | 48 (44.9±3.8) b | 46 (42.7 ± 3.4) b |
| 4 μM | 93 | 92 (99.2 ± 0.8) | 61 (66.6 ± 3.4) b | 24 (26.8±3.4) b | 24 (26.8±3.4) b | 20 (22.1 ± 1.8) c |
| 8 μM | 99 | 95 (96.0 ± 2.5) | 21 (21.4 ± 0.8) c | 0 (0) c | 0 (0) c | 0 (0) d |
| Non-IVF ** | 80 | 79 (99.6 ± 0.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haraguchi, S.; Ikeda, M.; Akagi, S.; Hirao, Y. Dynamic Changes in pStat3 Are Involved in Meiotic Spindle Assembly in Mouse Oocytes. Int. J. Mol. Sci. 2020, 21, 1220. https://doi.org/10.3390/ijms21041220
Haraguchi S, Ikeda M, Akagi S, Hirao Y. Dynamic Changes in pStat3 Are Involved in Meiotic Spindle Assembly in Mouse Oocytes. International Journal of Molecular Sciences. 2020; 21(4):1220. https://doi.org/10.3390/ijms21041220
Chicago/Turabian StyleHaraguchi, Seiki, Mitsumi Ikeda, Satoshi Akagi, and Yuji Hirao. 2020. "Dynamic Changes in pStat3 Are Involved in Meiotic Spindle Assembly in Mouse Oocytes" International Journal of Molecular Sciences 21, no. 4: 1220. https://doi.org/10.3390/ijms21041220
APA StyleHaraguchi, S., Ikeda, M., Akagi, S., & Hirao, Y. (2020). Dynamic Changes in pStat3 Are Involved in Meiotic Spindle Assembly in Mouse Oocytes. International Journal of Molecular Sciences, 21(4), 1220. https://doi.org/10.3390/ijms21041220




