Comparative Proteomic Analysis of Wild-Type Physcomitrella Patens and an OPDA-Deficient Physcomitrella Patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding
Abstract
:1. Introduction
2. Results and Discussion
2.1. Generation of 12-Oxo-phytodienoic acid (OPDA)-deficient P. Patens Mutants with Disrupted PpAOS1 and PpAOS2 Genes
2.2. Identification of Proteins that are Differentially Accumulated in Response to Wounding
2.3. Functional Categories of Identified Wounding-Responsive Proteins
2.4. Subcellular Localization of the Identified Proteins in Response to Wounding
2.5. Protein Catalogs of the Identified Proteins
2.5.1. Proteins Involved in Protein Synthesis
2.5.2. Proteins Involved in Protein Degradation
2.5.3. Proteins Involved in Amino Acid Metabolism
2.5.4. Proteins Involved in Protein Folding
2.5.5. Proteins Involved in Photosystems
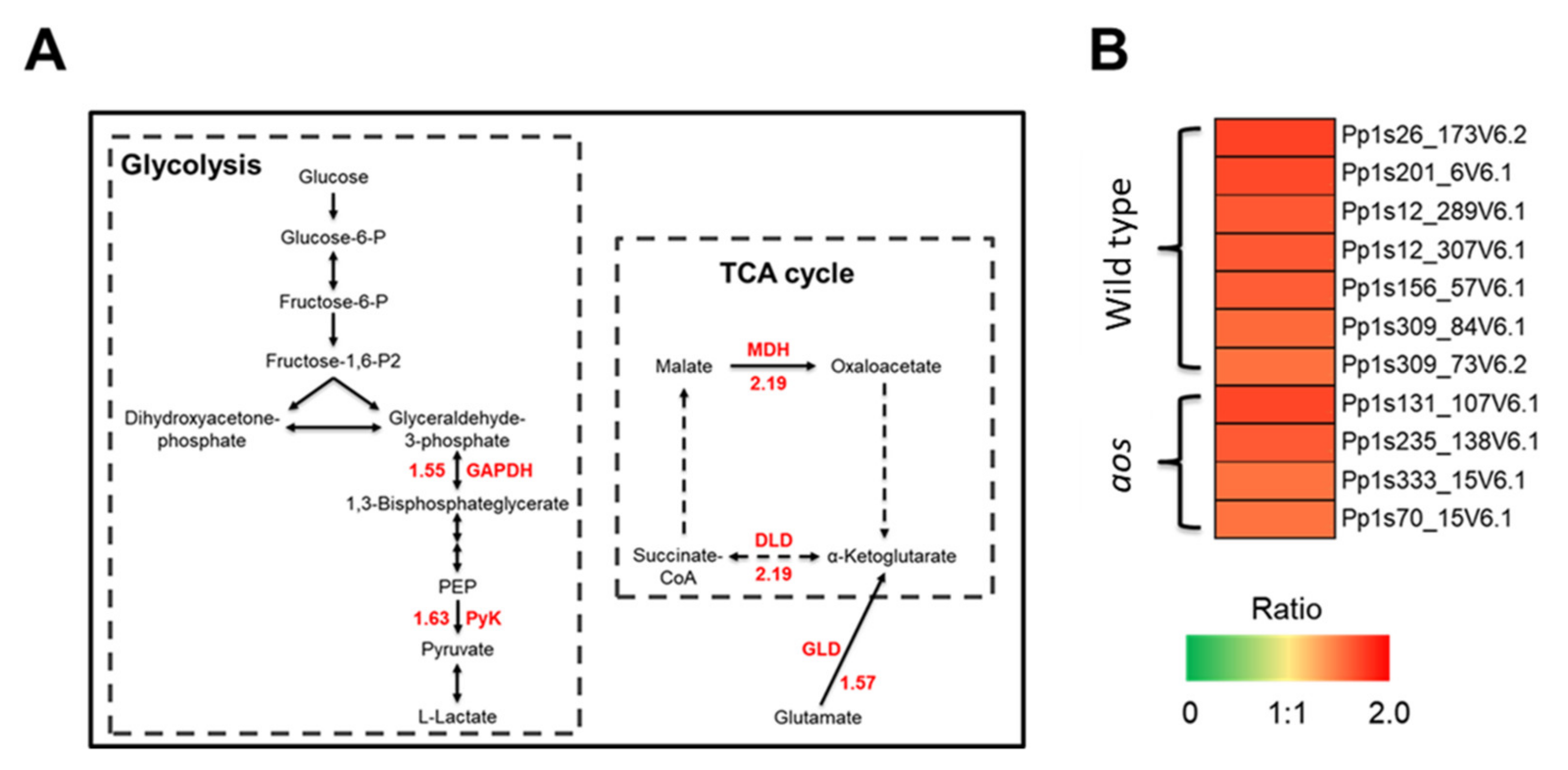
2.5.6. Proteins Involved in Glycolysis, the TCA Cycle, and Energy Synthesis
2.5.7. Proteins for Reactive Oxygen Scavenging
2.6. Quantitative RT-PCR Analysis of Genes Encoding Proteins Accumulated by Wounding
2.7. Comparison of Proteomic Data in This Study with Those in P. Patens Treated with OPDA
3. Materials and Methods
3.1. Plant Growth Conditions and Treatment
3.2. Analysis of OPDA Concentration in P. Patens
3.3. Generation of a P. Patens Mutant with Disrupted PpAOS1 and PpAOS2
3.4. Protein Extraction
3.5. Digestion of Proteins
3.6. Nanoliquid Chromatography-Tandem MS Analysis
3.7. Protein Identification Using Mascot
3.8. Analysis of Differentially Accumulated Proteins using the Acquired MS Data
3.9. Classification of Proteins and Bioinformatic Analysis
3.10. Quantitative RT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AOS | Allene oxide synthase |
| COI1 | Coronatine insensitive 1 |
| 12,13-EOT | 12,13(S)-epoxyoctadecatrienoic acid |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GLD | Glutamate dehydrogenase |
| GO | Gene Ontology |
| HPL | Hydroperoxide lyase |
| 13-HPOT | 13(S)-hydroperoxyoctadecatrienoic acid |
| HSP | Heat shock protein |
| JA | Jasmonic acid |
| JA-Ile | Isoleucine conjugate of jasmonic acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
| LEA | Late embryogenesis abundant |
| MDH | Malate dehydrogenase; |
| MeJA | Methyl jasmonate |
| OPDA | 12-Oxo-phytodienoic acid |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| ROS | Reactive oxygen species |
| Rubisco | Ribulose-1,5-bisphosphate carboxylase/oxygenase |
| STRING | Search Tool or the Retrieval of Interacting Genes |
| UPLC-MS/MS | Ultra-performance liquid chromatography-tandem mass spectrometry |
References
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [Green Version]
- León, J.; Rojo, E.; Sánchez-Serrano, J.J. Wound signalling in plants. J. Exp. Bot. 2001, 52, 1–9. [Google Scholar] [CrossRef]
- Nascimento, N.C.; Fett-Neto, A.G. Plant secondary metabolism and challenges in modifying its operation: an overview. Methods Mol. Biol. 2010, 643, 1–13. [Google Scholar] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Pratiwi, P.; Tanaka, G.; Takahashi, T.; Xie, X.; Yoneyama, K.; Matsuura, H.; Takahashi, K. Identification of jasmonic acid and jasmonoyl-isoleucine, and characterization of AOS, AOC, OPR and JAR1 in the model lycophyte Selaginella moellendorffii. Plant Cell Physiol. 2017, 58, 789–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browse, J.; Howe, G.A. New weapons and a rapid response against insect attack. Plant Physiol. 2008, 146, 832–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Ann. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Santino, A.; Taurino, M.; De Domenico, S.; Bonsegna, S.; Poltronieri, P.; Pastor, V.; Flors, V. Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 2013, 32, 1085–1098. [Google Scholar] [CrossRef]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, C.; Pollmann, S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009, 276, 4693–4704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumpe, M.; Göbel, C.; Faltin, B.; Beike, A.K.; Hause, B.; Himmelsbach, K.; Bode, J.; Kramell, R.; Wasternack, C.; Frank, W.; et al. The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 2010, 188, 740–749. [Google Scholar] [CrossRef]
- Anterola, A.; Göbel, C.; Hornung, E.; Sellhorn, G.; Feussner, I.; Grimes, H. Physcomitrella patens has lipoxygenases for both eicosanoid and octadecanoid pathways. Phytochemisty 2009, 70, 40–52. [Google Scholar] [CrossRef]
- Costa, C.L.; Arruda, P.; Benedetti, C.E. An Arabidopsis gene induced by wounding functionally homologous to flavoprotein oxidoreductases. Plant Mol. Biol. 2000, 44, 61–71. [Google Scholar] [CrossRef]
- Buseman, C.M.; Tamura, P.; Sparks, A.A.; Baughman, E.J.; Maatta, S.; Zhao, J.; Roth, M.R.; Esch, S.W.; Shah, J.; Williams, T.D.; et al. Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 2006, 142, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef] [Green Version]
- Dave, A.; Hernandez, M.L.; He, Z.; Andriotis, V.M.E.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 2011, 23, 583–599. [Google Scholar] [CrossRef] [Green Version]
- Dave, A.; Vaistij, F.E.; Gilday, A.D.; Penfield, S.D.; Graham, I.A. Regulation of Arabidopsis thaliana seed dormancy and germination by 12-oxo-phytodienoic acid. J. Exp. Bot. 2016, 67, 2277–2284. [Google Scholar] [CrossRef] [Green Version]
- Goetz, S.; Hellwege, A.; Stenzel, I.; Kutter, C.; Hauptmann, V.; Forner, S.; McCaig, B.; Hause, G.; Miersch, O.; Wasternack, C.; et al. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 2012, 158, 1715–1727. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.L.; Li, L.; Wang, B.; Chen, Z.; Knoop, V.; Groth-Malonek, M.; Dombrovska, O.; Lee, J.; Kent, L.; Rest, J.; et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl. Acad. Sci. USA 2006, 103, 15511–15516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.L.; Floyd, S.K.; Sakakibara, K. Green genes-comparative genomics of the green branch of life. Cell 2007, 129, 229–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.W.; Roberts, E.M.; Haigler, C.H. Moss cell walls: Structure and biosynthesis. Front. Plant Sci. 2012, 3, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yang, P.; Gao, Q.; Liu, X.; Kuang, T.; Shen, S.; He, Y. Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 2008, 228, 167–177. [Google Scholar] [CrossRef]
- Wang, X.; Yang, P.; Zhang, X.; Xu, Y.; Kuang, T.; Shen, S.; He, Y. Proteomic analysis of the cold stress response in the moss, Physcomitrella patens. Proteomics 2009, 9, 4529–4538. [Google Scholar] [CrossRef]
- Wang, X.; Kuang, T.; He, Y. Conservation between higher plants and the moss Physcomitrella patens in response to the phytohormone abscisic acid: A proteomics analysis. BMC Plant Biol. 2010, 10, 192. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, Y.; Yang, P. Proteomic studies of the abiotic stresses response in model moss—Physcomitrella patens. Front. Plant Sci. 2012, 3, 258. [Google Scholar] [CrossRef] [Green Version]
- Ponce de Leon, I.; Schmelz, E.A.; Gaggero, C.; Castro, A.; Álvarez, A.; Montesano, M. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 2012, 13, 960–974. [Google Scholar] [CrossRef]
- Toshima, E.; Nanjo, Y.; Komatsu, S.; Abe, T.; Matsuura, H.; Takahashi, K. Proteomic analysis of Physcomitrella patens treated with 12-oxo-phytodienoic acid, an important oxylipin in plants. Biosci. Biotechnol. Biochem. 2014, 78, 946–953. [Google Scholar] [CrossRef]
- Scholz, J.; Brodhun, F.; Hornung, E.; Herrfurth, C.; Stumpe, M.; Beike, A.K.; Faltin, B.; Frank, W.; Reski, R.; Feussner, I. Biosynthesis of allene oxides in Physcomitrella patens. BMC Plant Biol. 2012, 12, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosova, K.; Vitamvas, P.; Prasil, I.T.; Renaut, J. Plant proteome changes under abiotic stress–contribution of proteomics studies to understanding plant stress response. J. Proteomics 2011, 74, 1301–1322. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Proteomic analysis of calcium effects on soybean root tip under flooding and drought stresses. Plant Cell Physiol. 2017, 58, 1405–1420. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savatin, D.V.; Gramegna, G.; Modesti, V.; Cervone, F. Wounding in the plant tissues: The defense of a dangerous passage. Front. Plant Sci. 2014, 5, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2014, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Gruber, A.V.; Nisemblat, S.; Azem, A.; Weiss, C. The complexity of chloroplast chaperonins. Trends Plant Sci. 2013, 18, 688–694. [Google Scholar] [CrossRef]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2014, 9, 13–17. [Google Scholar] [CrossRef]
- Luo, W.; Nanjo, Y.; Komatsu, S.; Matsuura, H.; Takahashi, K. Proteomics of Physcomitrella patens protonemata subjected to treatment with 12-oxo-phytodienoic acid. Biosci. Biotechnol. Biochem. 2016, 80, 2357–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Hiwatashi, Y.; Sakakibara, K.; Kato, M.; Hasebe, M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 2000, 7, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, C.; Seto, Y.; Nabeta, K.; Matsuura, H. Kinetics of the accumulation of jasmonic acid and its derivatives in systemic leaves of tobacco (Nicotiana tabacum cv. Xanthi nc) and translocation of deuterium-labeled jasmonic acid from the wounding site to the systemic site. Biosci. Biotechol. Biochem. 2009, 73, 1962–1970. [Google Scholar] [CrossRef]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef]
- Nanjo, Y.; Skultety, L.; Uváčková, L.; Klubicová, K.; Hajduch, M.; Komatsu, S. Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteome Res. 2012, 11, 372–385. [Google Scholar] [CrossRef]
- Olsen, J.V.; de Godoy, L.M.F.; Li, G.; Macek, B.; Mortensen, P.; Pesch, R.; Makarov, A.; Lange, O.; Horning, S.; Mann, M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics 2005, 4, 2010–2021. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wen, Z.; Washburn, M.P.; Florens, L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 2009, 81, 6317–6326. [Google Scholar] [CrossRef]
- Brosch, M.; Yu, L.; Hubbard, T.; Choudhary, J. Accurate and sensitive peptide identification with Mascot Percolator. J. Proteome Res. 2009, 8, 3176–3181. [Google Scholar] [CrossRef] [Green Version]
- Bevan, M.; Bancroft, I.; Bent, E. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucl. Acids Res. 2011, 39 . [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucl. Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, L.; Yang, G.; Zhu, H.; He, Y. Transcriptome of protoplasts reprogrammed into stem cells in Physcomitrella patens. PLoS ONE 2012, 7, e35961. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Cortes, T.; Pomar, F.; Merino, F.; Novo-Uzai, E. A proteomic approach to Physcomitrella patens rhizoid exudates. J. Plant Physiol. 2014, 171, 1671–1678. [Google Scholar] [CrossRef]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, W.; Komatsu, S.; Abe, T.; Matsuura, H.; Takahashi, K. Comparative Proteomic Analysis of Wild-Type Physcomitrella Patens and an OPDA-Deficient Physcomitrella Patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding. Int. J. Mol. Sci. 2020, 21, 1417. https://doi.org/10.3390/ijms21041417
Luo W, Komatsu S, Abe T, Matsuura H, Takahashi K. Comparative Proteomic Analysis of Wild-Type Physcomitrella Patens and an OPDA-Deficient Physcomitrella Patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding. International Journal of Molecular Sciences. 2020; 21(4):1417. https://doi.org/10.3390/ijms21041417
Chicago/Turabian StyleLuo, Weifeng, Setsuko Komatsu, Tatsuya Abe, Hideyuki Matsuura, and Kosaku Takahashi. 2020. "Comparative Proteomic Analysis of Wild-Type Physcomitrella Patens and an OPDA-Deficient Physcomitrella Patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding" International Journal of Molecular Sciences 21, no. 4: 1417. https://doi.org/10.3390/ijms21041417
APA StyleLuo, W., Komatsu, S., Abe, T., Matsuura, H., & Takahashi, K. (2020). Comparative Proteomic Analysis of Wild-Type Physcomitrella Patens and an OPDA-Deficient Physcomitrella Patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding. International Journal of Molecular Sciences, 21(4), 1417. https://doi.org/10.3390/ijms21041417





