In Vivo Allergen-Activated Eosinophils Promote Collagen I and Fibronectin Gene Expression in Airway Smooth Muscle Cells via TGF-β1 Signaling Pathway in Asthma
Abstract
:1. Introduction
2. Results
2.1. Characteristics of the Study Population
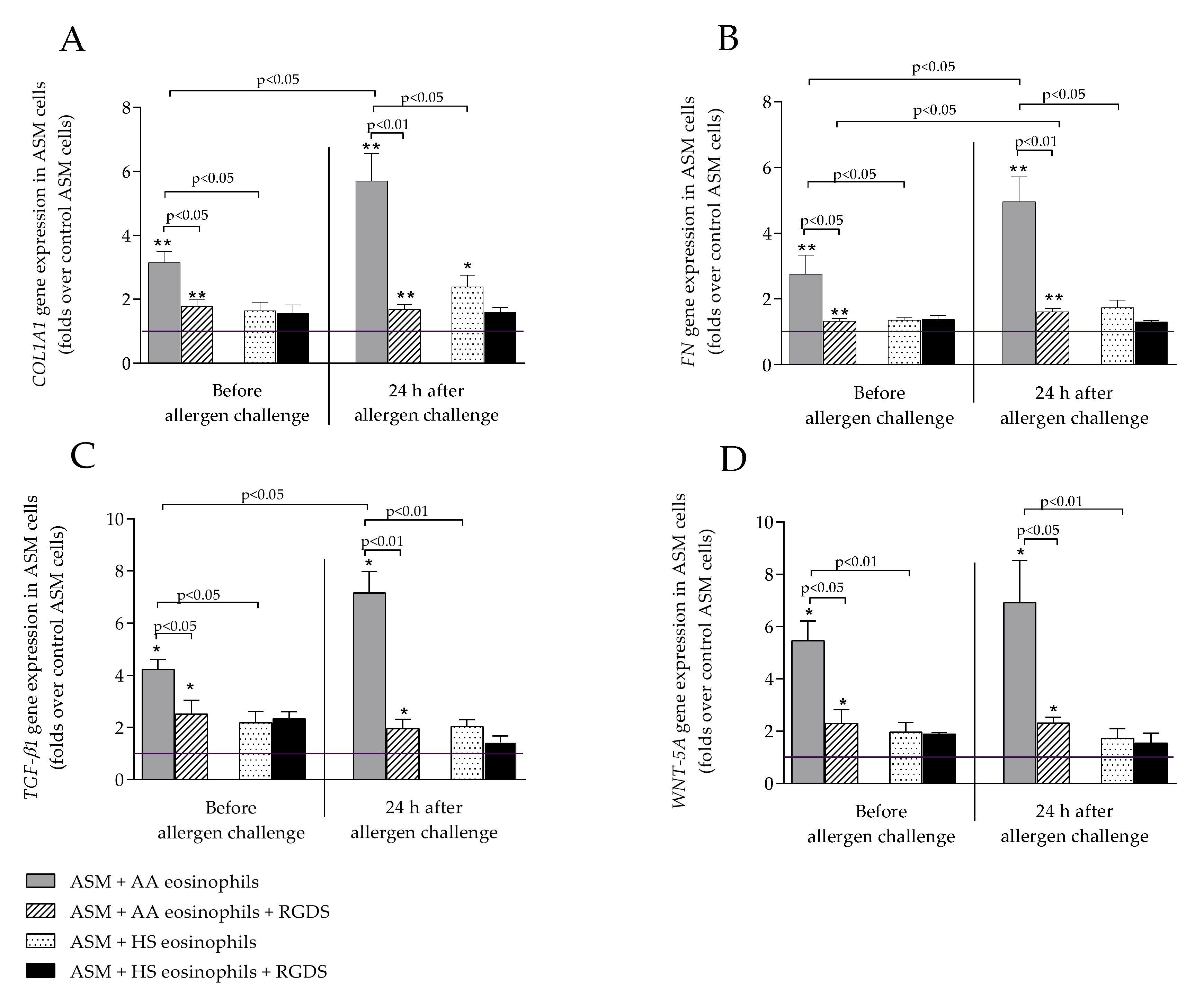
2.2. TGF-β1 Expression in Eosinophils and Airway Smooth Muscle Cells
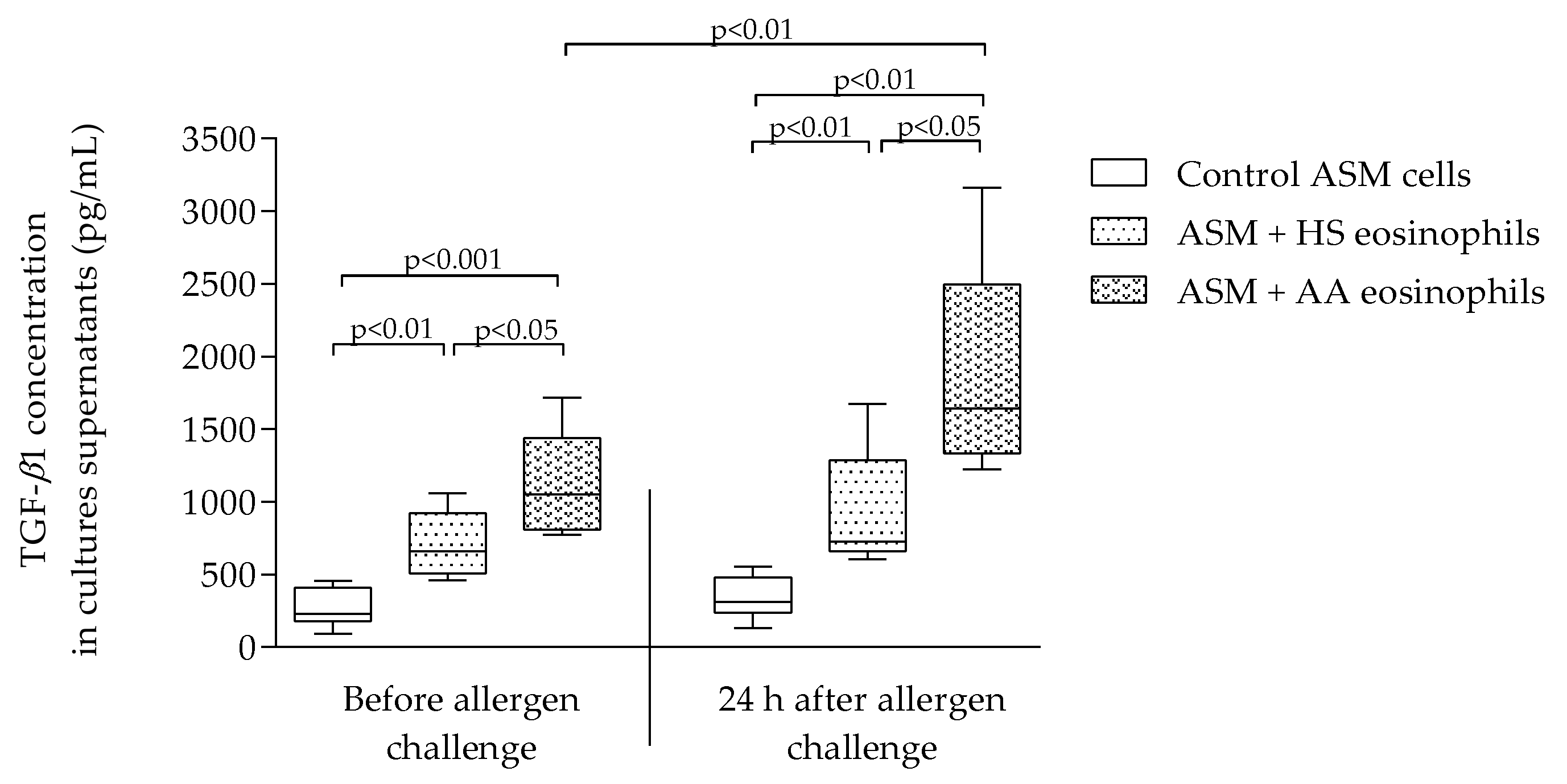
2.3. TGF-β1 Concentration in Culture Supernatants
2.4. WNT-5A Expression in Airway Smooth Muscle Cells
2.5. COL1A1 and FN Expression in Airway Smooth Muscle Cells
2.6. Suppression of Eosinophil Integrins with RGDS Peptide
3. Discussion
4. Materials and Methods
4.1. Study Subjects
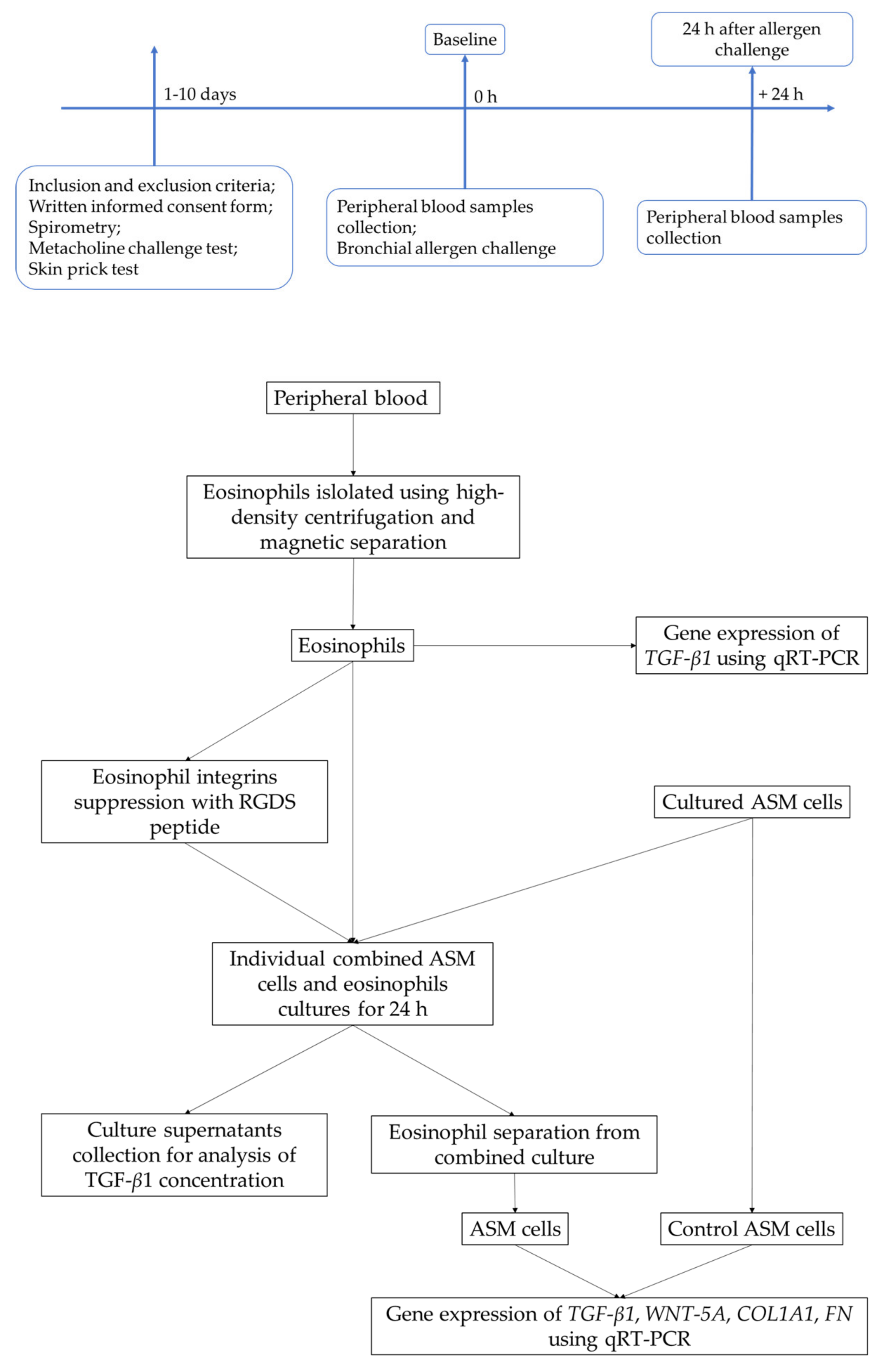
4.2. Study Design
4.3. Lung Function Testing
4.4. Measurement of Airway Responsiveness to Methacholine
4.5. Skin Prick Test
4.6. Bronchial Allergen Challenge
4.7. Isolation of Eosinophils from Peripheral Blood
4.8. Airway Smooth Muscle Cell Culture
4.9. Combined Culture of Airway Smooth Muscle Cells and Eosinophils
4.10. RNA Isolation and Quantitive Real-Time PCR Analysis
4.11. The Concentration of TGF-β1 in Culture Supernatants Analysis
4.12. Sputum Induction, Processing, and Cell Analysis
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | Allergic asthma |
| ASM | Airway smooth muscle |
| BMI | Body mass index |
| CCR3 | CC chemokine receptor 3 |
| COL1A1 | Collagen I alpha 1 gene |
| CTGF | Connective tissue growth factor |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DTT | Dithiothreitol |
| ECM | Extracellular matrix |
| ELISA | Enzyme-linked immunosorbent assay |
| ERK | Extracellular signal-regulated kinases |
| FBS | Fetal bovine serum |
| FEV1 | Forced expiratory volume in one second |
| FN | Fibronectin gene |
| FVC | Forced vital capacity |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| HS | Healthy subjects |
| hTERT | Human telomerase reverse transcriptase |
| IL | Interleukin |
| IR | Index of reactivity |
| ITS | Insulin-Transferrin-Selenium |
| JNK | c-Jun N-terminal kinases |
| K2EDTA | Ethylenediaminetetraacetic acid |
| MAPK | Mitogen-activated protein kinases |
| mRNA | Messenger ribonucleic acid |
| MMP | Matrix metalloproteinase |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| PBS | Phosphate buffer saline |
| PD20A | The provocative dose of allergen causing a 20% drop in FEV1 |
| PD20M | The provocative dose of methacholine causing a 20% drop in FEV1 |
| RGDS | Arginyl-glycyl-aspartyl-serine peptide (Arg-Gly-Asp-Ser) |
| ROS | Reactive oxygen species |
| RNA | Ribonucleic acid |
| SD | Standard deviation |
| SEM | Standard error of the mean |
| TGF-β1 | Transforming growth factor β1 gene |
| TGF-β1 | Transforming growth factor β1 |
| TSLP | Thymic stromal lymphopoietin |
| WNT-5A | Wingless integrase-1 5A gene |
| WNT-5A | Wingless integrase-1 5A |
References
- Kay, A.B.; Phipps, S.; Robinson, D.S. A role for eosinophils in airway remodelling in asthma. Trends Immunol. 2004, 25, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Fahy, J.V. Type 2 inflammation in asthma--present in most, absent in many. Nat. Rev. Immunol. 2015, 15, 57–65. [Google Scholar] [CrossRef] [PubMed]
- O’Byrne, P.M.; Inman, M.D. Airway Hyperresponsiveness. Chest 2003, 123, 411S–416S. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Ishigatsubo, Y.; Aoki, I. Pathology of asthma. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4. [Google Scholar] [CrossRef]
- Johansson, M.W. Eosinophil Activation Status in Separate Compartments and Association with Asthma. Front. Med. 2017, 4. [Google Scholar] [CrossRef]
- Bousquet, J.; Jeffery, P.K.; Busse, W.W.; Johnson, M.; Vignola, A.M. Asthma. From bronchoconstriction to airways inflammation and remodeling. Am. J. Respir. Crit. Care Med. 2000, 161, 1720–1745. [Google Scholar] [CrossRef] [Green Version]
- Ito, J.T.; Lourenço, J.D.; Righetti, R.F.; Tibério, I.F.L.C.; Prado, C.M.; Lopes, F.D.T.Q.S. Extracellular Matrix Component Remodeling in Respiratory Diseases: What Has Been Found in Clinical and Experimental Studies? Cells 2019, 8, 342. [Google Scholar] [CrossRef] [Green Version]
- Bara, I.; Ozier, A.; Tunon de Lara, J.M.; Marthan, R.; Berger, P. Pathophysiology of bronchial smooth muscle remodelling in asthma. Eur. Respir. J. 2010, 36, 1174–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuyderduyn, S.; Sukkar, M.B.; Fust, A.; Dhaliwal, S.; Burgess, J.K. Treating asthma means treating airway smooth muscle cells. Eur. Respir. J. 2008, 32, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, K.; Willems-Widyastuti, A.; Alagappan, V.K.T.; Radford, K.; Kranenburg, A.R.; Sharma, H.S. Role of extracellular matrix and its regulators in human airway smooth muscle biology. Cell Biochem. Biophys. 2006, 44, 139–146. [Google Scholar] [CrossRef]
- Al-Alawi, M.; Hassan, T.; Chotirmall, S.H. Transforming growth factor β and severe asthma: A perfect storm. Respir. Med. 2014, 108, 1409–1423. [Google Scholar] [CrossRef] [Green Version]
- Li, M.O.; Wan, Y.Y.; Sanjabi, S.; Robertson, A.-K.L.; Flavell, R.A. TRANSFORMING GROWTH FACTOR-β REGULATION OF IMMUNE RESPONSES. Annu. Rev. Immunol. 2006, 24, 99–146. [Google Scholar] [CrossRef] [PubMed]
- Oenema, T.A.; Mensink, G.; Smedinga, L.; Halayko, A.J.; Zaagsma, J.; Meurs, H.; Gosens, R.; Dekkers, B.G.J. Cross-Talk between Transforming Growth Factor–β1 and Muscarinic M2 Receptors Augments Airway Smooth Muscle Proliferation. Am. J. Respir. Cell Mol. Biol. 2013, 49, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Khalil, N.; Xu, Y.D.; O’Connor, R.; Duronio, V. Proliferation of Pulmonary Interstitial Fibroblasts Is Mediated by Transforming Growth Factor-β1-induced Release of Extracellular Fibroblast Growth Factor-2 and Phosphorylation of p38 MAPK and JNK. J. Biol. Chem. 2005, 280, 43000–43009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, H.; Cambier, S.; Somanath, S.; Barker, T.; Minagawa, S.; Markovics, J.; Goodsell, A.; Publicover, J.; Reichardt, L.; Jablons, D.; et al. Mouse and human lung fibroblasts regulate dendritic cell trafficking, airway inflammation, and fibrosis through integrin αvβ8–mediated activation of TGF-β. J. Clin. Investig. 2011, 121, 2863–2875. [Google Scholar] [CrossRef] [Green Version]
- Duvernelle, C.; Freund, V.; Frossard, N. Transforming growth factor-β and its role in asthma. Pulm. Pharmacol. Ther. 2003, 16, 181–196. [Google Scholar] [CrossRef]
- Kehrl, J.H.; Roberts, A.B.; Wakefield, L.M.; Jakowlew, S.; Sporn, M.B.; Fauci, A.S. Transforming growth factor beta is an important immunomodulatory protein for human B lymphocytes. J. Immunol. 1986, 137, 3855–3860. [Google Scholar]
- Oh, S.A.; Li, M.O. TGF-β: Guardian of T cell function. J. Immunol. 2013, 191, 3973–3979. [Google Scholar] [CrossRef]
- Ashcroft, G.S. Bidirectional regulation of macrophage function by TGF-beta. Microbes Infect. 1999, 1, 1275–1282. [Google Scholar] [CrossRef] [Green Version]
- Saito, A.; Horie, M.; Nagase, T. TGF-β Signaling in Lung Health and Disease. Int. J. Mol. Sci. 2018, 19, 2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januskevicius, A.; Vaitkiene, S.; Gosens, R.; Janulaityte, I.; Hoppenot, D.; Sakalauskas, R.; Malakauskas, K. Eosinophils enhance WNT-5a and TGF-beta1 genes expression in airway smooth muscle cells and promote their proliferation by increased extracellular matrix proteins production in asthma. Bmc Pulm. Med. 2016, 16, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Sjostrand, M.; Malmhall, C.; Radinger, M.; Jeurink, P.; Lotvall, J.; Bossios, A. New production of eosinophils and the corresponding TH1/TH2 balance in the lungs after allergen exposure in BALB/c and C57BL/6 mice. Scand. J. Immunol. 2010, 71, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Kouro, T.; Takatsu, K. IL-5- and eosinophil-mediated inflammation: From discovery to therapy. Int. Immunol. 2009, 21, 1303–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, H.; Takatsu, K. Role of cytokines in allergic airway inflammation. Int. Arch. Allergy Immunol. 2007, 142, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.D.; O’Byrne, P.M. Biologics and the lung: TSLP and other epithelial cell-derived cytokines in asthma. Pharmacol. Ther. 2017, 169, 104–112. [Google Scholar] [CrossRef]
- Kalinauskaite-Zukauske, V.; Januskevicius, A.; Janulaityte, I.; Miliauskas, S.; Malakauskas, K. Expression of eosinophil β chain-signaling cytokines receptors, outer-membrane integrins, and type 2 inflammation biomarkers in severe non-allergic eosinophilic asthma. Bmc Pulm. Med. 2019, 19, 158. [Google Scholar] [CrossRef] [Green Version]
- Calhoun, W.J.; Bates, M.E.; Schrader, L.; Sedgwick, J.B.; Busse, W.W. Characteristics of peripheral blood eosinophils in patients with nocturnal asthma. Am. Rev. Respir. Dis. 1992, 145, 577–581. [Google Scholar] [CrossRef]
- Wardlaw, A.J. Molecular basis for selective eosinophil trafficking in asthma: A multistep paradigm. J. Allergy Clin. Immunol. 1999, 104, 917–926. [Google Scholar] [CrossRef]
- Fulkerson, P.C.; Fischetti, C.A.; McBride, M.L.; Hassman, L.M.; Hogan, S.P.; Rothenberg, M.E. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc. Natl. Acad. Sci. USA 2006, 103, 16418–16423. [Google Scholar] [CrossRef] [Green Version]
- Barthel, S.R.; Johansson, M.W.; McNamee, D.M.; Mosher, D.F. Roles of integrin activation in eosinophil function and the eosinophilic inflammation of asthma. J. Leukoc. Biol. 2008, 83, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januskevicius, A.; Gosens, R.; Sakalauskas, R.; Vaitkiene, S.; Janulaityte, I.; Halayko, A.J.; Hoppenot, D.; Malakauskas, K. Suppression of Eosinophil Integrins Prevents Remodeling of Airway Smooth Muscle in Asthma. Front. Physiol. 2017, 7, 680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januskevicius, A.; Janulaityte, I.; Kalinauskaite-Zukauske, V.; Gosens, R.; Malakauskas, K. The Enhanced Adhesion of Eosinophils Is Associated with Their Prolonged Viability and Pro-Proliferative Effect in Asthma. J. Clin. Med. 2019, 8, 1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonbol, H.S. Extracellular Matrix Remodeling in Human Disease. J. Microsc. Ultrastruct. 2018, 6, 123–128. [Google Scholar] [CrossRef]
- Parameswaran, K.; Radford, K.; Zuo, J.; Janssen, L.J.; O’Byrne, P.M.; Cox, P.G. Extracellular matrix regulates human airway smooth muscle cell migration. Eur. Respir. J. 2004, 24, 545–551. [Google Scholar] [CrossRef]
- Somaiah, C.; Kumar, A.; Mawrie, D.; Sharma, A.; Patil, S.D.; Bhattacharyya, J.; Swaminathan, R.; Jaganathan, B.G. Collagen Promotes Higher Adhesion, Survival and Proliferation of Mesenchymal Stem Cells. PLoS ONE 2015, 10, e0145068. [Google Scholar] [CrossRef] [Green Version]
- Gerthoffer, W.T.; Schaafsma, D.; Sharma, P.; Ghavami, S.; Halayko, A.J. Motility, survival, and proliferation. Compr. Physiol. 2012, 2, 255–281. [Google Scholar] [CrossRef] [Green Version]
- Hirst, S.J.; Twort, C.H.; Lee, T.H. Differential effects of extracellular matrix proteins on human airway smooth muscle cell proliferation and phenotype. Am. J. Respir. Cell Mol. Biol. 2000, 23, 335–344. [Google Scholar] [CrossRef] [Green Version]
- Freyer, A.M.; Johnson, S.R.; Hall, I.P. Effects of growth factors and extracellular matrix on survival of human airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 2001, 25, 569–576. [Google Scholar] [CrossRef]
- Koohestani, F.; Braundmeier, A.G.; Mahdian, A.; Seo, J.; Bi, J.; Nowak, R.A. Extracellular matrix collagen alters cell proliferation and cell cycle progression of human uterine leiomyoma smooth muscle cells. PLoS ONE 2013, 8, e75844. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.R.; Burgess, J.K.; Underwood, P.A.; Au, W.; Poniris, M.H.; Tamm, M.; Ge, Q.; Roth, M.; Black, J.L. Extracellular matrix proteins modulate asthmatic airway smooth muscle cell proliferation via an autocrine mechanism. J. Allergy Clin. Immunol. 2004, 113, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Chan, V.; Burgess, J.K.; Ratoff, J.C.; O’Connor, B.J.; Greenough, A.; Lee, T.H.; Hirst, S.J. Extracellular Matrix Regulates Enhanced Eotaxin Expression in Asthmatic Airway Smooth Muscle Cells. Am. J. Respir. Crit. Care Med. 2006, 174, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Araujo, B.B.; Dolhnikoff, M.; Silva, L.F.F.; Elliot, J.; Lindeman, J.H.N.; Ferreira, D.S.; Mulder, A.; Gomes, H.A.P.; Fernezlian, S.M.; James, A.; et al. Extracellular matrix components and regulators in the airway smooth muscle in asthma. Eur. Respir. J. 2008, 32, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minshall, E.M.; Leung, D.Y.; Martin, R.J.; Song, Y.L.; Cameron, L.; Ernst, P.; Hamid, Q. Eosinophil-associated TGF-beta1 mRNA expression and airways fibrosis in bronchial asthma. Am. J. Respir. Cell Mol. Biol. 1997, 17, 326–333. [Google Scholar] [CrossRef]
- Pascoe, C.D.; Obeidat, M.e.; Arsenault, B.A.; Nie, Y.; Warner, S.; Stefanowicz, D.; Wadsworth, S.J.; Hirota, J.A.; Jasemine Yang, S.; Dorscheid, D.R.; et al. Gene expression analysis in asthma using a targeted multiplex array. Bmc Pulm. Med. 2017, 17, 189. [Google Scholar] [CrossRef]
- Johnson, P.R.A.; Burgess, J.K.; Ge, Q.; Poniris, M.; Boustany, S.; Twigg, S.M.; Black, J.L. Connective Tissue Growth Factor Induces Extracellular Matrix in Asthmatic Airway Smooth Muscle. Am. J. Respir. Crit. Care Med. 2006, 173, 32–41. [Google Scholar] [CrossRef]
- Ahmadzai, M.; Small, M.; Sehmi, R.; Gauvreau, G.; Janssen, L.J. Integrins are Mechanosensors That Modulate Human Eosinophil Activation. Front. Immunol. 2015, 6, 525. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Aparicio, P.; Dominguez-Jiménez, C.; Garcia-Pardo, A. Activation of the alpha 4 beta 1 integrin through the beta 1 subunit induces recognition of the RGDS sequence in fibronectin. J. Cell Biol. 1994, 126, 271–279. [Google Scholar] [CrossRef] [Green Version]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef]
- D’Souza, S.E.; Ginsberg, M.H.; Plow, E.F. Arginyl-glycyl-aspartic acid (RGD): A cell adhesion motif. Trends Biochem. Sci. 1991, 16, 246–250. [Google Scholar] [CrossRef]
- Pfaff, M. Recognition sites of RGD-dependent integrins. In Integrin-ligand interaction; Springer: Boston, MA, USA, 1997; pp. 101–121. [Google Scholar]
- Cook, N.S.; Kottirsch, G.; Zerwes, H.-G. Platelet glycoprotein IIb/IIIa antagonists. Drugs Future 1994, 19, 135–159. [Google Scholar]
- Verrecchia, F.; Mauviel, A. Transforming Growth Factor-β Signaling Through the Smad Pathway: Role in Extracellular Matrix Gene Expression and Regulation. J. Investig. Dermatol. 2002, 118, 211–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halwani, R.; Al-Muhsen, S.; Al-Jahdali, H.; Hamid, Q. Role of Transforming Growth Factor–β in Airway Remodeling in Asthma. Am. J. Respir. Cell Mol. Biol. 2011, 44, 127–133. [Google Scholar] [CrossRef]
- Xiao, Y.Q.; Freire-de-Lima, C.G.; Schiemann, W.P.; Bratton, D.L.; Vandivier, R.W.; Henson, P.M. Transcriptional and Translational Regulation of TGF-β Production in Response to Apoptotic Cells. J. Immunol. 2008, 181, 3575–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badewa, A.P.; Heiman, A.S. Inhibition of CCL11, CCL24, and CCL26-induced degranulation in HL-60 eosinophilic cells by specific inhibitors of MEK1/MEK2, p38 MAP kinase, and PI 3-kinase. Immunopharmacol. Immunotoxicol. 2003, 25, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Gerthoffer, W.T.; Singer, C.A. MAPK regulation of gene expression in airway smooth muscle. Respir. Physiol. Neurobiol. 2003, 137, 237–250. [Google Scholar] [CrossRef]
- Lavinskiene, S.; Malakauskas, K.; Jeroch, J.; Hoppenot, D.; Sakalauskas, R. Functional activity of peripheral blood eosinophils in allergen-induced late-phase airway inflammation in asthma patients. J. Inflamm. 2015, 12, 25. [Google Scholar] [CrossRef] [Green Version]
- Schwingshackl, A.; Duszyk, M.; Brown, N.; Moqbel, R. Human eosinophils release matrix metalloproteinase-9 on stimulation with TNF-α. J. Allergy Clin. Immunol. 1999, 104, 983–990. [Google Scholar] [CrossRef]
- Ohno, I.; Ohtani, H.; Nitta, Y.; Suzuki, J.; Hoshi, H.; Honma, M.; Isoyama, S.; Tanno, Y.; Tamura, G.; Yamauchi, K.; et al. Eosinophils as a source of matrix metalloproteinase-9 in asthmatic airway inflammation. Am. J. Respir. Cell Mol. Biol. 1997, 16, 212–219. [Google Scholar] [CrossRef]
- Tatler, A.L.; John, A.E.; Jolly, L.; Habgood, A.; Porte, J.; Brightling, C.; Knox, A.J.; Pang, L.; Sheppard, D.; Huang, X.; et al. Integrin αvβ5-mediated TGF-β activation by airway smooth muscle cells in asthma. J Immunol. 2011, 187, 6094–6107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [Green Version]
- Ignotz, R.A.; Massague, J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J. Biol. Chem. 1986, 261, 4337–4345. [Google Scholar] [PubMed]
- Ojiaku, C.A.; Yoo, E.J.; Panettieri, R.A. Transforming Growth Factor β1 Function in Airway Remodeling and Hyperresponsiveness. The Missing Link? Am. J. Respir. Cell Mol. Biol. 2016, 56, 432–442. [Google Scholar] [CrossRef]
- McMillan, S.J.; Xanthou, G.; Lloyd, C.M. Manipulation of allergen-induced airway remodeling by treatment with anti-TGF-beta antibody: Effect on the Smad signaling pathway. J Immunol. 2005, 174, 5774–5780. [Google Scholar] [CrossRef] [Green Version]
- Manuyakorn, W.; Kamchaisatian, W.; Atamasirikul, K.; Sasisakulporn, C.; Direkwattanachai, C.; Benjaponpitak, S. Serum TGF-beta1 in atopic asthma. Asian Pac. J. Allergy Immunol. 2008, 26, 185–189. [Google Scholar]
- McKinley, L.; Alcorn, J.F.; Peterson, A.; Dupont, R.B.; Kapadia, S.; Logar, A.; Henry, A.; Irvin, C.G.; Piganelli, J.D.; Ray, A.; et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J. Immunol. 2008, 181, 4089–4097. [Google Scholar] [CrossRef] [Green Version]
- Wakashin, H.; Hirose, K.; Maezawa, Y.; Kagami, S.; Suto, A.; Watanabe, N.; Saito, Y.; Hatano, M.; Tokuhisa, T.; Iwakura, Y.; et al. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am. J. Respir. Crit. Care Med. 2008, 178, 1023–1032. [Google Scholar] [CrossRef]
- Bajoriuniene, I.; Malakauskas, K.; Lavinskiene, S.; Jeroch, J.; Sakalauskas, R. Th17 response to Dermatophagoides pteronyssinus is related to late-phase airway and systemic inflammation in allergic asthma. Int. Immunopharmacol. 2013, 17, 1020–1027. [Google Scholar] [CrossRef]
- Lan, F.; Liu, K.; Zhang, J.; Qi, Y.; Li, K.; Lin, P. Th17 response is augmented in OVA-induced asthmatic mice exposed to HDM. Med. Sci. Monit. 2011, 17, BR132–BR138. [Google Scholar] [CrossRef]
- Al-Muhsen, S.; Letuve, S.; Vazquez-Tello, A.; Pureza, M.A.; Al-Jahdali, H.; Bahammam, A.S.; Hamid, Q.; Halwani, R. Th17 cytokines induce pro-fibrotic cytokines release from human eosinophils. Respir Res 2013, 14, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, S.; Martin, H.; Beckert, H.; Bros, M.; Montermann, E.; Belz, C.; Heinz, A.; Ohngemach, S.; Sahin, U.; Stassen, M.; et al. The Wnt/beta-catenin pathway attenuates experimental allergic airway disease. J. Immunol. 2014, 193, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koopmans, T.; Hesse, L.; Nawijn, M.; Menzen, M.; Smits, R.; Bakker, E.; Vonk, J.; van Den Berge, M.; Koppelman, G.; Guryev, V.; et al. Smooth-muscle-derived WNT-5A drives allergen-induced remodelling and Th2 type inflammation. Eur. Respir. J. 2018, 52, PA5256. [Google Scholar] [CrossRef]
- Kumawat, K.; Menzen, M.H.; Bos, I.S.T.; Baarsma, H.A.; Borger, P.; Roth, M.; Tamm, M.; Halayko, A.J.; Simoons, M.; Prins, A.; et al. Noncanonical WNT-5A signaling regulates TGF-β-induced extracellular matrix production by airway smooth muscle cells. Faseb J. 2013, 27, 1631–1643. [Google Scholar] [CrossRef]
- Koopmans, T.; Gosens, R. Revisiting asthma therapeutics: Focus on WNT signal transduction. Drug Discov. Today 2018, 23, 49–62. [Google Scholar] [CrossRef]
- Burgess, J.K.; Oliver, B.G.; Poniris, M.H.; Ge, Q.; Boustany, S.; Cox, N.; Moir, L.M.; Johnson, P.R.; Black, J.L. A phosphodiesterase 4 inhibitor inhibits matrix protein deposition in airways in vitro. J. Allergy Clin. Immunol. 2006, 118, 649–657. [Google Scholar] [CrossRef]
- Wong, D.T.; Elovic, A.; Matossian, K.; Nagura, N.; McBride, J.; Chou, M.Y.; Gordon, J.R.; Rand, T.H.; Galli, S.J.; Weller, P.F. Eosinophils from patients with blood eosinophilia express transforming growth factor beta 1. Blood 1991, 78, 2702–2707. [Google Scholar] [CrossRef] [Green Version]
- Gauvreau, G.M.; Watson, R.M.; O’Byrne, P.M. Kinetics of allergen-induced airway eosinophilic cytokine production and airway inflammation. Am. J. Respir. Crit. Care Med. 1999, 160, 640–647. [Google Scholar] [CrossRef]
- Duncan, C.J.A.; Lawrie, A.; Blaylock, M.G.; Douglas, J.G.; Walsh, G.M. Reduced eosinophil apoptosis in induced sputum correlates with asthma severity. Eur. Respir. J. 2003, 22, 484–490. [Google Scholar] [CrossRef] [Green Version]
- Denlinger, L.C.; Phillips, B.R.; Ramratnam, S.; Ross, K.; Bhakta, N.R.; Cardet, J.C.; Castro, M.; Peters, S.P.; Phipatanakul, W.; Aujla, S.; et al. Inflammatory and Comorbid Features of Patients with Severe Asthma and Frequent Exacerbations. Am. J. Respir. Crit. Care Med. 2017, 195, 302–313. [Google Scholar] [CrossRef]
- Gosens, R.; Stelmack, G.L.; Dueck, G.; McNeill, K.D.; Yamasaki, A.; Gerthoffer, W.T.; Unruh, H.; Gounni, A.S.; Zaagsma, J.; Halayko, A.J. Role of caveolin-1 in p42/p44 MAP kinase activation and proliferation of human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 291, L523–L534. [Google Scholar] [CrossRef] [PubMed]

| AA Patients, n = 12 | HS, n = 11 | |||
|---|---|---|---|---|
| Age, median (range), years | 28.5 (20–44) | 26.0 (23–42) | ||
| Sex, (male/female), n | 4/8 | 5/6 | ||
| BMI, kg/m2 | 22.4 ± 2.6 | 24.0 ± 5.1 | ||
| Sensitization to D. pteronyssinus/D. farinae/birch/five grass mixture allergen, n | 12/11/2/4 | NR | ||
| Wheel diameter by D. pteronyssinus, median (range), mm | 5 (3–8) | NR | ||
| PD20M, geometric mean (range), mg | 0.09 (0.007–0.260) | NR | ||
| PD20A, geometric mean (range), IR/mL | 6.684 (1.631–9.403) | NR | ||
| Maximum fall in FEV1after bronchial allergen challenge, mean % (min–max) | –31.2 (−52.1–−22.4) | −3.8 (−7.2–0.0) | ||
| FEV1, % of predicted | 99.0 ± 5.73 | 102.0 ± 7.05 | ||
| FEV1, L | 3.69 ± 0.36 | 4.14 ± 0.54 | ||
| Baseline | 24 h after allergen challenge | Baseline | 24 h after allergen challenge | |
| Blood eosinophil count, ×109/L | 0.34 ± 0.11 * # | 0.52 ± 0.30 # | 0.15 ± 0.06 | 0.16 ± 0.04 |
| Blood eosinophil count, % | 7.08 ± 3.98 * # | 8.63 ± 3.01 # | 2.00 ± 1.05 | 2.51 ± 0.74 |
| Sputum cell viability, % (AA n = 9, HS n = 7) | 70.5 ± 5.34 * # | 79.9 ± 11.2 # | 51.5 ± 14.9 | 57.5 ± 12.3 |
| Sputum eosinophil count, % (AA n = 9, HS n = 7) | 5.5 ± 5.4 * # | 13.3 ± 12.87 # | 0.1 ± 0.2 | 0.5 ± 0.4 |
| Gene | Forward 5′-3′ | Reverse 5′-3′ |
|---|---|---|
| 18S | CGCCGCTAGAGGTGAAATTC | TTGGCAAATGCTTTCGCTC |
| WNT-5a | GGGTGGGAACCAAGAAAAAT | TGGAACCTACCCATCCCATA |
| TGF-β1 | GTACCTGAACCCGTGTTGCT | GAACCCGTTGATGTCCACTT |
| COL1A1 | TCGAGGAGGAAATTCCAATG | ACACACGTGCACCTCATCAT |
| FN | AGCCAGCAGATCGAGAACAT | TCTTGTCCTTGGGGTTCTTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janulaityte, I.; Januskevicius, A.; Kalinauskaite-Zukauske, V.; Bajoriuniene, I.; Malakauskas, K. In Vivo Allergen-Activated Eosinophils Promote Collagen I and Fibronectin Gene Expression in Airway Smooth Muscle Cells via TGF-β1 Signaling Pathway in Asthma. Int. J. Mol. Sci. 2020, 21, 1837. https://doi.org/10.3390/ijms21051837
Janulaityte I, Januskevicius A, Kalinauskaite-Zukauske V, Bajoriuniene I, Malakauskas K. In Vivo Allergen-Activated Eosinophils Promote Collagen I and Fibronectin Gene Expression in Airway Smooth Muscle Cells via TGF-β1 Signaling Pathway in Asthma. International Journal of Molecular Sciences. 2020; 21(5):1837. https://doi.org/10.3390/ijms21051837
Chicago/Turabian StyleJanulaityte, Ieva, Andrius Januskevicius, Virginija Kalinauskaite-Zukauske, Ieva Bajoriuniene, and Kestutis Malakauskas. 2020. "In Vivo Allergen-Activated Eosinophils Promote Collagen I and Fibronectin Gene Expression in Airway Smooth Muscle Cells via TGF-β1 Signaling Pathway in Asthma" International Journal of Molecular Sciences 21, no. 5: 1837. https://doi.org/10.3390/ijms21051837





