Phylogeny and Functions of LOB Domain Proteins in Plants
Abstract
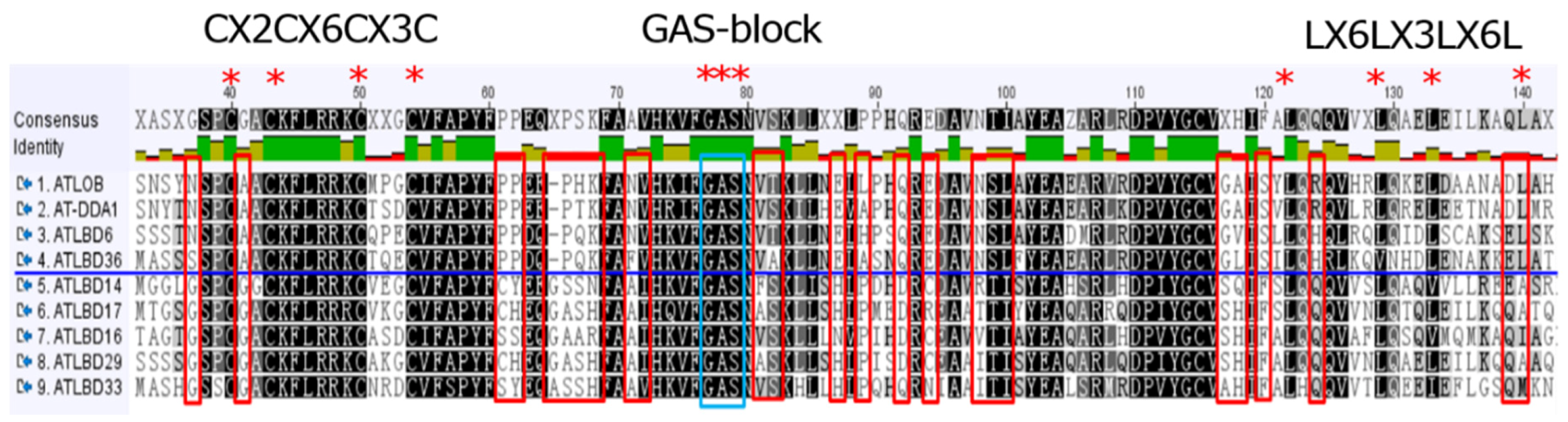
:1. The Confusion in Understanding Functional Diversity of LBD Proteins
2. The Phylogeny of LBD Family Proteins
3. Functional Conservation and Diversity of LBD Proteins in Arabidopsis
4. Function Summarization Under Phylogenetic Framework
4.1. Functions of LBD Proteins in Class IA Clade
4.2. Functions of LBD Proteins in Class IB Clade
4.3. Functions of LBD Proteins in Class IC Clade
4.4. Functions of LBD Proteins in Class IE Clade
4.5. Functions of LBD Proteins in Class II Clade
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, W.F.; Wei, X.-B.; Rety, S.; Huang, L.Y.; Liu, N.N.; Dou, S.X.; Xi, X.G. Structural analysis reveals a “molecular calipers” mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat. J. Biol. Chem. 2019, 294, 142–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The Lateral Organ Boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, M.J.; Park, M.Y.; Han, K.H.; Kim, J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant 2013, 6, 1722–1725. [Google Scholar] [CrossRef] [Green Version]
- Majer, C.; Xu, C.; Berendzen, K.W.; Hochholdinger, F. Molecular interactions of Rootless Concerning Crown and Seminal Roots, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1542–1551. [Google Scholar] [CrossRef] [Green Version]
- Kong, Y.; Xu, P.; Jing, X.; Chen, L.; Li, L.; Li, X. Decipher the ancestry of the plant-specific LBD gene family. BMC Genom. 2017, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chanderbali, A.S.; He, F.; Soltis, P.S.; Soltis, D.E. Out of the water: Origin and diversification of the LBD gene family. Mol. Biol. Evol. 2015, 32, 1996–2000. [Google Scholar] [CrossRef] [Green Version]
- Hochholdinger, F.; Zimmermann, R. Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 2008, 11, 70–74. [Google Scholar] [CrossRef]
- Yu, P.; Gutjahr, C.; Li, C.; Hochholdinger, F. Genetic control of lateral root formation in cereals. Trends Plant Sci. 2016, 21, 951–961. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Yu, P.; Marcon, C. Genetic control of root system development in. Trends Plant Sci. 2018, 23, 79–88. [Google Scholar] [CrossRef]
- Liu, W.; Yu, J.; Ge, Y.; Qin, P.; Xu, L. Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018, 75, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Hake, S. How a leaf gets its shape. Curr. Opin. Plant Biol. 2011, 14, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Kazan, K.; Manners, J.M. Lateral organ boundaries domain transcription factors: New roles in plant defense. Plant Signal. Behav. 2012, 7, 1702–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangeon, A.; Bell, E.M.; Lin, W.C.; Jablonska, B.; Springer, P.S. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot. 2011, 62, 221–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Moreau, C.; Liu, Y.; Kawaguchi, M.; Hofer, J.; Ellis, N.; Chen, R. Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc. Natl. Acad. Sci. USA 2012, 109, 11723–11728. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Han, L.; Fu, C.; Chai, M.; Zhang, W.; Li, G.; Tang, Y.; Wang, Z.Y. Identification and characterization of petiolule- like pulvinus mutants with abolished nyctinastic leaf movement in the model legume Medicago truncatula. New Phytol. 2012, 196, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Ge, L.; Peng, J.; Berbel, A.; Madueño, F.; Chen, R. Regulation of compound leaf development by PHANTASTICA in Medicago truncatula. Plant Physiol. 2014, 164, 216–228. [Google Scholar] [CrossRef] [Green Version]
- Thatcher, L.F.; Powell, J.J.; Aitken, E.A.B.; Kazan, K.; Manners, J.M. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, J.; Díaz-Manzano, F.E.; Sanchez, M.; Rosso, M.N.; Melillo, T.; Goh, T.; Fukaki, H.; Cabello, S.; Hofmann, J.; Fenoll, C.; et al. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014, 203, 632–645. [Google Scholar] [CrossRef]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Yamashino, T.; Kiba, T.; Koizumi, N.; Kojima, M.; Sakakibara, H.; Mizuno, T. A link between cytokinin and ASL9 (Asymmetric Leaves 2 Like 9) that belongs to the AS2/LOB (Lateral Organ Boundaries) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [Green Version]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Xiaofei, W.; Xin, L.; Ling, S.; Yongjiang, S.; Shizhong, Z.; Yujin, H.; Chunxiang, Y. Identification, Evolution and Expression Analysis of the LBD Gene Family in Tomato. Sci. Agric. Sin. 2013, 46, 2501–2513. [Google Scholar]
- Wang, X.; Zhang, S.; Su, L.; Liu, X.; Hao, Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE 2013, 8, e57044. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Liu, W.; Xie, W.; Liu, Z.; Liu, Z.; Wang, Y. Genome-wide Analysis of the LBD Transcription Factor Family in Medicago truncatula. Xibei Zhiwu Xuebao 2014, 34, 2176–2187. [Google Scholar]
- Zhang, Y.M.; Zhang, S.Z.; Zheng, C.C. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef]
- Cao, H.; Liu, C.Y.; Liu, C.X.; Zhao, Y.L.; Xu, R.R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016, 95, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Ma, B.; Zeng, Q.; Xiang, Z.; He, N. Identification and characterization of Lateral Organ Boundaries Domain genes in mulberry, Morus notabilis. Meta Gene 2016, 8, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.J.; Wang, J.; Lin, S.; Tian, Z.; Zhou, K.; Luan, H.Y.; Lyu, C.; Zhang, X.Z.; Xu, R.G. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ. Sci. B 2016, 17, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Chen, D.; Zheng, Y.; Jin, S.; Yang, J.; Ye, N. Identification and expression analyses of sbp-box genes reveal their involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 3404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Shao, F.; Macmillan, C.; Wilson, I.W.; van der Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Guo, A.Y.; Gao, G.; Zhong, Y.F.; Xu, M.; Huang, M.; Luo, J. DPTF: A database of poplar transcription factors. Bioinformatics 2007, 23, 1307–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coudert, Y.; Dievart, A.; Droc, G.; Gantet, P. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol. Biol. Evol. 2013, 30, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Taramino, G.; Sauer, M.; Stauffer, J.L.; Multani, D.; Niu, X.; Sakai, H.; Hochholdinger, F. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 2007, 50, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an Auxin Response Factor in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, T.; Joi, S.; Mimura, T.; Fukaki, H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 2012, 139, 883–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef] [Green Version]
- Cheng, P.C.; Greyson, R.I.; Walden, D.B. Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am. J. Bot. 1983, 70, 450. [Google Scholar] [CrossRef]
- Luo, J.H.; Weng, L.; Luo, D. Isolation and expression patterns of Lateral Organ Boundaries- like genes in Lotus japonicus. J. Plant Physiol. Mol. Biol. 2006, 32, 202–208. [Google Scholar]
- Wang, L.; Wang, W.; De-Yang, X.U.; Chang, Z.Y.; Cui, X.F. Overexpression of Arabidopsis ASL25/LBD28 Gene Affects Leaf Morphogenesis. Acta Bot. Boreali Occident. Sin. 2010, 30, 888–893. [Google Scholar]
- Ohashi-Ito, K.; Iwamoto, K.; Fukuda, H. LOB DOMAIN-CONTAINING PROTEIN 15 positively regulates expression of VND7, a master regulator of tracheary elements. Plant Cell Physiol. 2018, 59, 989–996. [Google Scholar] [CrossRef] [Green Version]
- Cho, C.; Jeon, E.; Pandey, S.K.; Ha, S.H.; Kim, J. LBD13 positively regulates lateral root formation in Arabidopsis. Planta 2019, 249, 1251–1258. [Google Scholar] [CrossRef]
- Koppolu, R.; Anwar, N.; Sakuma, S.; Tagiri, A.; Lundqvist, U.; Pourkheirandish, M.; Rutten, T.; Seiler, C.; Himmelbach, A.; Ariyadasa, R.; et al. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. USA 2013, 110, 13198–13203. [Google Scholar] [CrossRef] [Green Version]
- Jeon, B.W.; Kim, J. Role of LBD14 during ABA-mediated control of root system architecture in Arabidopsis. Plant Signal. Behav. 2018, 13, e1507405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Cao, H.; Zhang, Q.; Wang, H.; Xin, W.; Xu, E.; Zhang, S.; Yu, R.; Yu, D.; Hu, Y. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wu, F.; Sheng, P.; Wang, X.; Zhang, Z.; Zhou, K.; Zhang, H.; Hu, J.; Lin, Q.; Cheng, Z.; et al. The LBD12-1 transcription factor suppresses apical meristem size by repressing argonaute 10 expression. Plant Physiol. 2017, 173, 801–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Dai, Z.; Li, L.; Wang, J.; Miao, X.; Shi, Z. OsRAMOSA2 shapes panicle architecture through regulating pedicel length. Front. Plant Sci. 2017, 8, 1538. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zou, X.; Zhang, C.; Shao, Q.; Liu, J.; Liu, B.; Li, H.; Zhao, T. OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS ONE 2016, 11, e0156413. [Google Scholar] [CrossRef]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.M.S. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef] [Green Version]
- Ariel, F.; Diet, A.; Verdenaud, M.; Gruber, V.; Frugier, F.; Chan, R.; Crespi, M. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-zip i transcription factor HB1. Plant Cell 2010, 22, 2171–2183. [Google Scholar] [CrossRef] [Green Version]
- Gendron, J.M.; Liu, J.S.; Fan, M.; Bai, M.Y.; Wenkel, S.; Springer, P.S.; Barton, M.K.; Wang, Z.Y. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 21152–21157. [Google Scholar] [CrossRef] [Green Version]
- Bell, E.M.; Lin, W.C.; Husbands, A.Y.; Yu, L.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.M.; Girke, T.; Springer, P.S. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef] [Green Version]
- Husbands, A.; Bell, E.M.; Shuai, B.; Smith, H.M.S.; Springer, P.S. Lateral organ boundaries defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007, 35, 6663–6671. [Google Scholar] [CrossRef]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C.P. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.C.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-Domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wang, H.; Li, J.; Huang, H.; Xu, L. Quantitative control of ASYMMETRIC LEAVES2 expression is critical for leaf axial patterning in Arabidopsis. J. Exp. Bot. 2013, 64, 4895–4905. [Google Scholar] [CrossRef] [Green Version]
- Rast, M.I.; Simon, R. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 2012, 24, 2917–2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Kim, M.J.; Pandey, S.; Kim, J. Expression and protein interaction analyses reveal combinatorial interactions of LBD transcription factors during Arabidopsis pollen development. Plant Cell Physiol. 2016, 57, 2291–2299. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, M.; Kim, J. Combinatorial interactions between LBD10 and LBD27 are essential for male gametophyte development in Arabidopsis. Plant Signal. Behav. 2015, 10, 14–16. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Kim, M.; Lee, M.R.; Park, S.K.; Kim, J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 2015, 81, 794–809. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, F.; Guo, J.; Zhang, X.S. Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J. Plant Biol. 2009, 52, 374–381. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, W.; Huang, Y.; Niu, X.; Zhao, Y.; Han, Y.; Liu, Y. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J. Exp. Bot. 2015, 66, 99–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.J.; Tao, Y.; Zhao, X.Y.; Zhang, X.S. Wheat TaAS2, a member of LOB family, affects the adaxial-abaxial polarity of leaves in transgenic Arabidopsis. Plant Sci. 2007, 172, 181–188. [Google Scholar] [CrossRef]
- Jeon, E.; Kang, N.Y.; Cho, C.; Seo, P.J.; Suh, M.C.; Kim, J. LBD14/ASL17 positively regulates lateral root formation and is involved in ABA response for root architecture in Arabidopsis. Plant Cell Physiol. 2017, 58, 2190–2201. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef] [Green Version]
- Berckmans, B.; Vassileva, V.; Schmid, S.P.C.; Maes, S.; Parizot, B.; Naramoto, S.; Magyar, Z.; Lessa Alvim Kamei, C.; Koncz, C.; Bögre, L.; et al. Auxin-Dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 2011, 23, 3671–3683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soyano, T.; Thitamadee, S.; Machida, Y.; Chua, N.H. Asymmetric Leaves2-Like19/Lateral Organ Boundaries Domain30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 2008, 20, 3359–3373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Wang, B.; Li, X.; Pan, J.; Qian, X.; Yu, Y.; Xu, P.; Zhu, J.; Xu, X. Lateral Organ Boundaries Domain 19 (LBD19) negative regulate callus formation in Arabidopsis. Plant Cell. Tissue Organ Cult. 2019, 137, 485–494. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-wide identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol. 2018, 59, 744–755. [Google Scholar] [CrossRef]
- Borghi, L.; Bureau, M.; Simon, R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 2007, 19, 1795–1808. [Google Scholar] [CrossRef] [Green Version]
- Rast-Somssich, M.I.; Zádníková, P.; Schmid, S.; Kieffer, M.; Kepinski, S.; Simon, R. The Arabidopsis JAGGED LATERAL ORGANS (JLO) gene sensitizes plants to auxin. J. Exp. Bot. 2017, 68, 2741–2755. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Zhang, Y.; Wu, X.; Tang, W.; Wu, R.; Dai, Z.; Liu, G.; Zhang, H.; Wu, C.; Chen, G.; et al. DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol. Biol. 2008, 66, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Coudert, Y.; Le, V.A.T.; Adam, H.; Bès, M.; Vignols, F.; Jouannic, S.; Guiderdoni, E.; Gantet, P. Identification of CROWN ROOTLESS1-regulated genes in rice reveals specific and conserved elements of postembryonic root formation. New Phytol. 2015, 206, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coudert, Y.; Bès, M.; Van Anh Le, T.; Pré, M.; Guiderdoni, E.; Gantet, P. Transcript profiling of crown rootless1 mutant stem base reveals new elements associated with crown root development in rice. BMC Genom. 2011, 12, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, S.; Yu, X.; Yu, J.; He, X.; Zhang, S.; Shou, H.; Wu, P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Muthreich, N.; Majer, C.; Beatty, M.; Paschold, A.; Schützenmeister, A.; Fu, Y.; Malik, W.A.; Schnable, P.S.; Piepho, H.P.; Sakai, H.; et al. Comparative transcriptome profiling of maize coleoptilar nodes during shoot-borne root initiation. Plant Physiol. 2013, 163, 419–430. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, M.; Ichikawa, T.; Ishikawa, A.; Kobayashi, H.; Tsuhara, Y.; Kawashima, M.; Suzuki, K.; Muto, S.; Matsui, M. Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 2003, 34, 741–750. [Google Scholar] [CrossRef]
- Sun, X.D.; Feng, Z.H.; Meng, L.S.; Zhu, J.; Geitmann, A. Arabidopsis ASL11/LBD15 is involved in shoot apical meristem development and regulates WUS expression. Planta 2013, 237, 1367–1378. [Google Scholar] [CrossRef]
- Wang, Y.B.; Song, J.P.; Wang, Z.B. ASYMMETRIC LEAVES2-LIKE38, one member of AS2/LOB gene family, involves in regulating ab-adaxial patterning in Arabidopsis lateral organs. Acta Physiol. Plant. 2015, 37, 185. [Google Scholar] [CrossRef]
- Erik, V.; Springer, P.S.; Lindee, G.; Buckler, E.S.; Robert, M. Architecture of floral branch systems in maize and related grasses. Nature 2005, 436, 1119–1126. [Google Scholar]
- Cortizo, M.; Laufs, P. Genetic basis of the “sleeping leaves” revealed. Proc. Natl. Acad. Sci.USA 2012, 109, 11474–11475. [Google Scholar] [CrossRef] [Green Version]
- Bureau, M.; Rast, M.I.; Illmer, J.; Simon, R. JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol. Biol. 2010, 74, 479–491. [Google Scholar] [CrossRef]
- Bureau, M.; Simon, R. JLO regulates embryo patterning and organ initiation by controlling auxin transport. Plant Signal. Behav. 2008, 3, 145–147. [Google Scholar] [CrossRef] [Green Version]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.J.; Park, J.W.; Lee, H.W.; Kim, J. Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J. Exp. Bot. 2009, 60, 3935–3957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Sun, X.; Wang, G.; Liu, H.; Zhu, J. LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann. Bot. 2012, 110, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porco, S.; Larrieu, A.; Du, Y.; Gaudinier, A.; Goh, T.; Swarup, K.; Swarup, R.; Kuempers, B.; Bishopp, A.; Lavenus, J.; et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 2016, 143, 3340–3349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auguy, F.; Laplaze, L.; Parizot, B.; Baker, A.; Ricaud, L.; Martinie, A.; Franche, C.; Nussaume, L.; Bogusz, D.; Haseloff, J. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 2433–2442. [Google Scholar]
- Lee, H.W.; Cho, C.; Kim, J. Lateral organ boundaries domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 1792–1806. [Google Scholar] [CrossRef] [Green Version]
- Marchant, A.; Bhalerao, R.; Casimiro, I.; Eklöf, J.; Casero, P.J.; Bennett, M.; Sandberg, G. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 2002, 14, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Swarup, K.; Benková, E.; Swarup, R.; Casimiro, I.; Péret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef]
- Pandey, S.K.; Lee, H.W.; Kim, M.J.; Cho, C.; Oh, E.; Kim, J. LBD18 uses a dual mode of a positive feedback loop to regulate ARF expression and transcriptional activity in Arabidopsis. Plant J. 2018, 95, 233–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Kim, J. Coiled-coil motif in LBD16 and LBD18 transcription factors are critical for dimerization and biological function in Arabidopsis. Plant Signal. Behav. 2018, 13, e1411450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis Regeneration from Multiple Tissues Occurs via a Root Development Pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hu, X.; Qin, P.; Prasad, K.; Hu, Y.; Xu, L. The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 2018, 59, 734–743. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, J.; Fenoll, C.; Escobar, C. Genes co-regulated with LBD16 in nematode feeding sites inferred from in silico analysis show similarities to regulatory circuits mediated by the auxin/cytokinin balance in Arabidopsis. Plant Signal. Behav. 2015, 10, 16–19. [Google Scholar] [CrossRef] [Green Version]
- Gil, J.F.; Liebe, S.; Thiel, H.; Lennfors, B.L.; Kraft, T.; Gilmer, D.; Maiss, E.; Varrelmann, M.; Savenkov, E.I. Massive up-regulation of LBD transcription factors and EXPANSINs highlights the regulatory programs of rhizomania disease. Mol. Plant Pathol. 2018, 19, 2333–2348. [Google Scholar]
- Oh, S.A.; Park, K.S.; Twell, D.; Park, S.K. The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J. 2010, 64, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Twell, D.; Park, S.K. SIDECAR POLLEN suggests a plant-specific regulatory network underlying asymmetric microspore division in Arabidopsis. Plant Signal. Behav. 2011, 6, 416–419. [Google Scholar] [CrossRef] [Green Version]
- Scheible, W.R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [Green Version]
- Graeff, M.; Straub, D.; Eguen, T.; Dolde, U.; Rodrigues, V.; Brandt, R.; Wenkel, S. MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 2016, 12, e1005959. [Google Scholar] [CrossRef] [Green Version]







| General Classification | Species | Gene Numbers in LBD Family | Data Source | Gene ID Prefix |
|---|---|---|---|---|
| Green alga | Cylindrocystis brebissonii | 6 | China National GeneBank Database | RPGL/YOXI |
| Moss | Physcomitrella patens | 30 | Gramene Database | Pp |
| Fern | Selaginella moellendorffii | 24 | Gramene Database | SELMO |
| Basal angiosperm | Amborella trichopoda | 20 | Gramene Database | AMTR |
| Eudicots | Arabidopsis thaliana | 43 | Gramene Database | AT |
| Beta vulgaris | 23 | Gramene Database | BVRB | |
| Brassica napus | 118 | Gramene Database | Bna | |
| Daucus carota | 54 | Gramene Database | DCAR | |
| Glycine max | 79 | Gramene Database | GLYMA | |
| Gossypium raimondii | 65 | Gramene Database | B456 | |
| Medicago truncatula | 57 | Gramene Database | MTR | |
| Solanum tuberosum | 39 | Gramene Database | PGSC | |
| Monocots | Hordeum vulgare | 28 | Gramene Database | HORVU |
| Oryza sativa | 37 | Gramene Database | BGIOSGA | |
| Setaria italica | 32 | Gramene Database | SETIT | |
| Sorghum bicolor | 33 | Gramene Database | SORBI | |
| Triticum aestivum | 86 | Gramene Database | TraesCS | |
| Zea mays | 49 | Gramene Database | Zm00001d |
| Classes | Species | Genes | Biological Processes | Target or Downstream Genes | References |
|---|---|---|---|---|---|
| Class IA | Arabidopsis | AtLBD25/ DDA1 | Auxin signaling, photomorphogenesis | - (no reports so far) | [15] |
| Arabidopsis | AtLOB/ ASL4 | Leaf adaxial–abaxial polarity, floral organ development, brassinasteroid accumulation | BAS1 | [2,59,60,61] | |
| Arabidopsis | AtLBD6/AS2 | Leaf adaxial–abaxial polarity, floral organ development | BP, KNAT2 | [62,63,64,65,66] | |
| Arabidopsis | AtLBD36/AS1 | ||||
| Arabidopsis | AtLBD10 | Pollen development | - | [67,68,69] | |
| Arabidopsis | AtLBD28 | Leaf adaxial–abaxial polarity, SAM genesis | - | [47] | |
| Rice | OsLBD1-9/ OsAS2; OsIG1 | Shoot differentiation, leaf development | - | [70] | |
| Empty-glume identity, floral organ number control, female gametophyte development | EG1, OsMADS1 and OsMADS6 | [71] | |||
| Rice | OsRAMOSA2/ OsRA2 | Panicle architecture establishment | LAX1, RCN2 | [54] | |
| Maize | ramosa2/ ra2 | Inflorescence architecture establishment | - | [56] | |
| Maize | IG1 | Embryo sac, leaf and tassel development | KN1, RS1, LG3, LG4A/B, and KNOX3 | [57] | |
| Wheat | TaAS2 | Leaf adaxial–abaxial polarity | - | [72] | |
| Barley | Vrs4/ HvRA2 | Control lateral spikelet fertility and row-type pathway | Vrs1, HvSRA and T6P | [50] | |
| Apple | MdLBD11 | Leaf and flower development | - | [28] | |
| Lotus japonicus | LjLOB1 | Compound leaf development, Floral development | - | [46] | |
| LjLOB3 | |||||
| Lotus japonicus | SLP | Pulvinus development, leaf movement | - | [16] | |
| Medicago | ELP1/ PLP | Pulvinus development, leaf movement | - | [16,17,18] | |
| Pea | APU | Pulvinus development, leaf movement | - | [16] | |
| Class IB | Arabidopsis | AtLBD14 | ABA-mediated lateral roots formation | AtHB7 and AtHB12 | [51,73] |
| Arabidopsis | AtLBD16 | Lateral roots formation, callus formation, gall formation | FAD-BD, ARF7 and ARF19 | [21,40,43,44,52,74] | |
| Arabidopsis | AtLBD17 | Auxin-induced callus formation | - | [52,74] | |
| Arabidopsis | AtLBD18 | Tracheary element differentiation, lateral roots formation, callus formation, auxin signaling | ARF7 and ARF19, EXP14, E2Fa, VND6 and VND7 | [44,74,75,76] | |
| Arabidopsis | AtLBD19 | Callus formation | - | [77] | |
| Arabidopsis | AtLBD20 | Fusarium wilt susceptibility, jasmonate signaling | MYC2, Thi2.1, VSP2, and PDF1.2 | [19] | |
| Arabidopsis | AtLBD29 | Lateral roots formation, callus formation, auxin signaling | ARF7 and ARF19 | [40,52,74,78] | |
| Arabidopsis | AtLBD30/ JLO | Tracheary element differentiation, embryogenesis, shoot, root and floral meristems development, auxin signaling | VND6 and VND7 | [66,76,79,80] | |
| Arabidopsis | AtLBD33 | Auxin-mediated lateral roots formation | ARF7 and ARF19, E2Fa | [75] | |
| Rice | OsDH1 | Floral organ development | - | [81] | |
| Rice | Crownrootless1 (crl1); OsARL1 | Auxin-mediated crown root formation | FSM, GTE4 and MAP | [42,82,83] | |
| Auxin-mediated adventitious root formation | - | [84] | |||
| Maize | ZmLBD2/ RTCS | Embryonic seminal and post-embryonic shoot-borne root initiation, auxin signaling | ZmArf34 | [5,41,85] | |
| Maize | ZmLBD43/ RTCL | ||||
| Lotus japonicus | LjLOB4 | Compound leaf development, Floral development | - | [46] | |
| Class IC | Arabidopsis | AtLBD3/ ASL9 | Cytokinin-responsive biological events | - | [23] |
| Arabidopsis | AtLBD12 | Growth of abaxial leaf surface, apical dominance and fertility | - | [86] | |
| Arabidopsis | AtLBD13 | Lateral root formation | - | [49] | |
| Arabidopsis | AtLBD15 | Tracheary element development, SAM development | WUS | [48,87] | |
| Rice | OsLBD3-7 | Leaf rolling | - | [55] | |
| Rice | OsLBD12-1 | SAM size control | AGO10 | [53] | |
| Eucalyptus grandis | EgLBD29 | Secondary xylem formation, response to Gibberellin (GA) and indol-3-acetic acid (IAA) | - | [35] | |
| Eucalyptus grandis | EgLBD37 | Secondary phloem formation, response to GA and IAA | - | [35] | |
| Citrus | CsLOB1 | Citrus bacterial canker disease susceptibility | - | [20] | |
| Class IE | Arabidopsis | AtLBD27 | Pollen development | - | [67,68,69] |
| Class II | Arabidopsis | AtLBD40 | GA response | - | [22] |
| Arabidopsis | AtLBD41/ ASL38 | Leaf adaxial-abaxial polarity | - | [88] | |
| Arabidopsis | AtLBD37 | Anthocyanin synthesis, nitrogen responses | PAP1 and PAP2 | [24] | |
| Arabidopsis | AtLBD38 | ||||
| Arabidopsis | AtLBD39 | ||||
| Rice | OsLBD37 | Nitrogen metabolism, heading date | Ehd1 | [25,26] | |
| Rice | OsLBD38 | Heading date | Ehd1 | [26] | |
| Medicago | MtLBD1 | Lateral roots emergence, auxin signaling | HB1 | [58] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Z.; Ma, B.; Hou, Q.; Wan, X. Phylogeny and Functions of LOB Domain Proteins in Plants. Int. J. Mol. Sci. 2020, 21, 2278. https://doi.org/10.3390/ijms21072278
Zhang Y, Li Z, Ma B, Hou Q, Wan X. Phylogeny and Functions of LOB Domain Proteins in Plants. International Journal of Molecular Sciences. 2020; 21(7):2278. https://doi.org/10.3390/ijms21072278
Chicago/Turabian StyleZhang, Yuwen, Ziwen Li, Biao Ma, Quancan Hou, and Xiangyuan Wan. 2020. "Phylogeny and Functions of LOB Domain Proteins in Plants" International Journal of Molecular Sciences 21, no. 7: 2278. https://doi.org/10.3390/ijms21072278






