Protein Synthesis by Day 16 Bovine Conceptuses during the Time of Maternal Recognition of Pregnancy
Abstract
:1. Introduction
2. Results
2.1. Experiment 1
2.1.1. Proteins Present in Conceptus-Conditioned Media following 6 h Culture of Day 16 Conceptuses in Vitro
2.1.2. De novo Proteins Synthesized by Day 16 Conceptuses following Culture in Vitro
2.2. Experiment 2
2.2.1. Specific Proteins are Unique to Extracellular Vesicles Recovered from Uterine Luminal Fluid of Pregnant Heifers on Day 16
2.2.2. Comparison between Conceptus-Derived Proteins and Those Present in the EVs from ULF
2.3. Comparison with Previous Studies
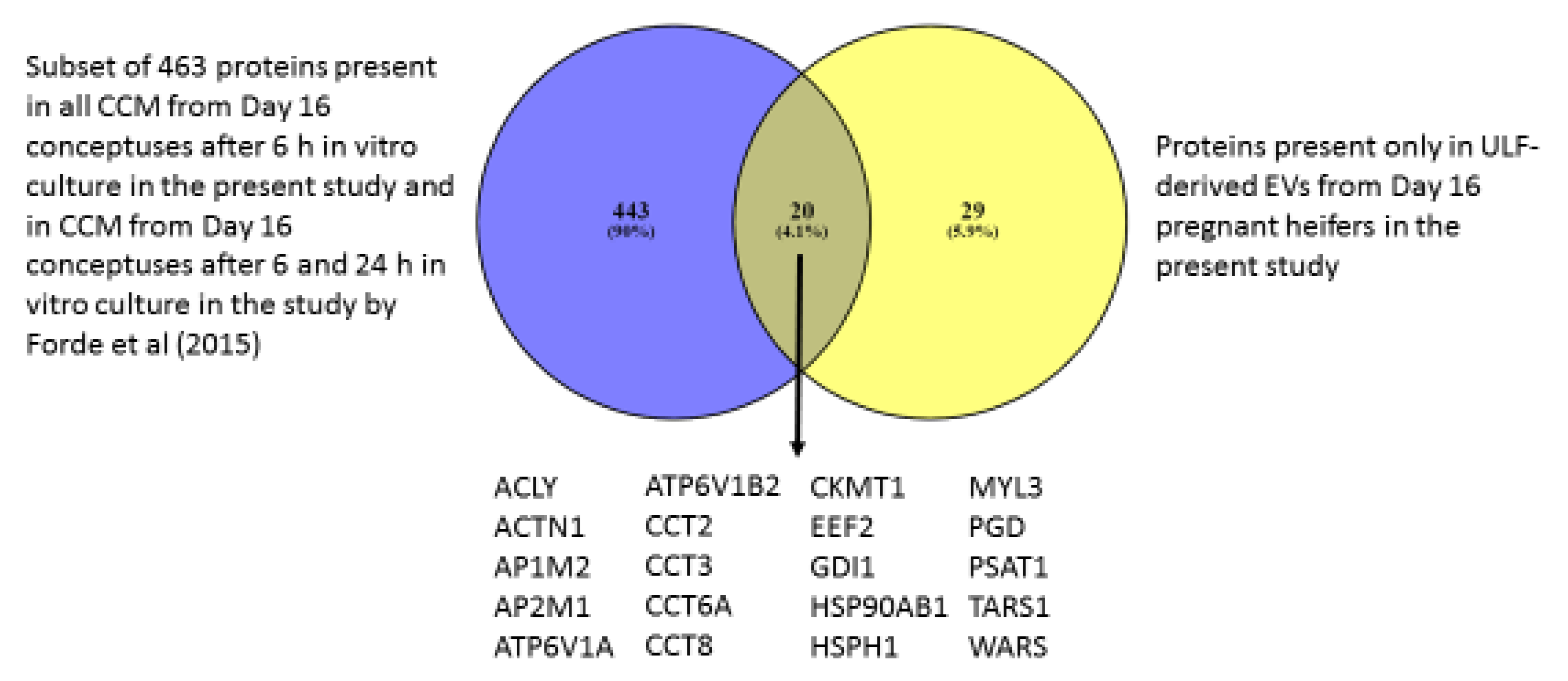
2.3.1. Comparison between Conceptus-Derived Proteins and Previously Published Work on CCM
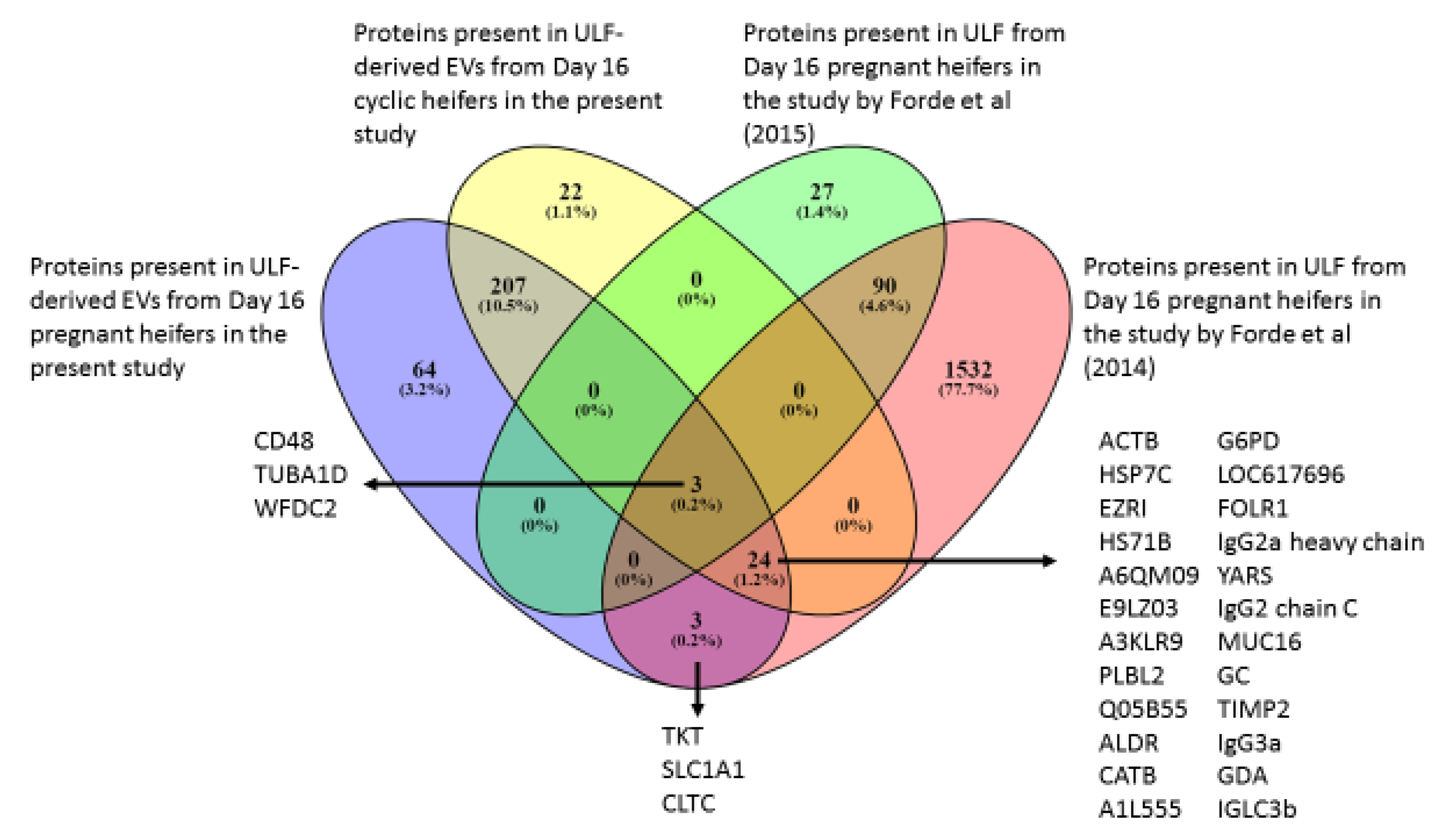
2.3.2. Comparison of EV Components to Protein Composition of ULF from Previously Published Studies
3. Discussion
3.1. De novo Synthesised Proteins in Conceptus-Conditioned Media
3.2. Proteins Detected in the ULF-Derived EVs
3.3. Comparison of EVs from ULF vs. CCM Proteins to Determine Those Potentially Involved in IFNT-Independent Communication during the Peri-Implantation Period of Pregnancy in Cattle
3.4. CCM vs. Previously Published Studies
4. Materials and Methods
4.1. Experiment 1: Identification of de novo Synthesised Proteins Produced by Day 16 Conceptuses Following Short-Term Culture In Vitro
4.1.1. Animal Synchronization and Sample Collection
4.1.2. Analysis of Protein Content of Conceptus-Conditioned Medium (CCM)
4.1.3. Data Analysis
4.2. Experiment 2: Proteomic Component of Extracellular Vesicles Obtained from the Uterine Luminal Fluid of Cyclic and Pregnant Heifers on Day 16
4.2.1. Animal Synchronization and Sample Collection
4.2.2. Extraction of Extracellular Vesicles from Uterine Luminal Fluid and Proteomic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maillo, V.; Gaora, P.Ó.; Forde, N.; Besenfelder, U.; Havlicek, V.; Burns, G.W.; Spencer, T.E.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Oviduct-Embryo Interactions in Cattle: Two-Way Traffic or a One-Way Street? Biol. Reprod. 2015, 92, 144. [Google Scholar] [CrossRef] [PubMed]
- Rizos, D.; Maillo, V.; Lonergan, P. Role of the oviduct and oviduct-derived products in ruminant embryo development. Anim. Reprod. 2016, 13, 160–167. [Google Scholar] [CrossRef]
- Cordova, A.; Perreau, C.; Uzbekova, S.; Ponsart, C.; Locatelli, Y.; Mermillod, P. Development rate and gene expression of IVP bovine embryos cocultured with bovine oviduct epithelial cells at early or late stage of preimplantation development. Theriogenology 2014, 81, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Rizos, D.; Ward, F.; Duffy PA, T.; Boland, M.P.; Lonergan, P. Consequences of bovine oocyte maturation, fertilization or early embryo development In Vitro versus In Vivo: Implications for blastocyst yield and blastocyst quality. Mol. Reprod. Dev. 2002, 61, 234–248. [Google Scholar] [CrossRef]
- Sponchiado, M.; Gomes, N.S.; Fontes, P.K.; Martins, T.; del Collado, M.; de Assumpcao Pastore, A.; Pugliesi, G.; Nogueira, M.F.G.; Binelli, M. Pre-hatching embryo-dependent and -independent programming of endometrial function in cattle. PLoS ONE 2017, 12, e0175954. [Google Scholar] [CrossRef] [PubMed]
- Passaro, C.; Tutt, D.; Mathew, D.J.; Sanchez, J.M.; Browne, J.A.; Boe-Hansen, G.B.; Fair, T.; Lonergan, P. Blastocyst-induced changes in the bovine endometrial transcriptome. Reproduction 2018, 156, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Passaro, C.; Tutt, D.; Bagés-Arnal, S.; Maicas, C.; Laguna-Barraza, R.; Gutierrez-Adan, A.; Browne, J.A.; Rath, D.; Behura, S.K.; Spencer, T.E.; et al. Global transcriptomic response of bovine endometrium to blastocyst-stage embryos. Reproduction 2019, 158, 223–235. [Google Scholar] [CrossRef]
- Spencer, T.E.; Forde, N.; Dorniak, P.; Hansen, T.R.; Romero, J.J.; Lonergan, P. Conceptus-derived prostaglandins regulate gene expression in the endometrium prior to pregnancy recognition in ruminants. Reproduction 2013, 146, 377–387. [Google Scholar] [CrossRef] [Green Version]
- Forde, N.; Duffy, G.B.; McGettigan, P.A.; Browne, J.A.; Mehta, J.P.; Kelly, A.K.; Mansouri-Attia, N.; Sandra, O.; Loftus, B.J.; Crowe, M.A.; et al. Evidence for an early endometrial response to pregnancy in cattle: Both dependent upon and independent of interferon tau. Physiol. Genom. 2012, 44, 799–810. [Google Scholar] [CrossRef]
- Forde, N.; Carter, F.; Spencer, T.E.; Bazer, F.W.; Sandra, O.; Mansouri-Attia, N.; Okumu, L.A.; McGettigan, P.A.; Mehta, J.P.; McBride, R.; et al. Conceptus-induced changes in the endometrial transcriptome: How soon does the cow know she is pregnant? Biol. Reprod. 2011, 85, 144–156. [Google Scholar] [CrossRef] [Green Version]
- Bauersachs, S.; Ulbrich, S.E.; Gross, K.; Schmidt, S.E.M.; Meyer, H.H.D.; Wenigerkind, H.; Vermehren, M.; Sinowatz, F.; Blum, H.; Wolf, E. Embryo-induced transcriptome changes in bovine endometrium reveal species-specific and common molecular markers of uterine receptivity. Reproduction 2006, 132, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauersachs, S.; Ulbrich, S.E.; Reichenbach, H.D.; Reichenbach, M.; Büttner, M.; Meyer, H.H.D.; Spencer, T.E.; Minten, M.; Sax, G.; Winter, G.; et al. Comparison of the Effects of Early Pregnancy with Human Interferon, Alpha 2 (IFNA2), on Gene Expression in Bovine Endometrium. Biol. Reprod. 2012, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gifford, C.A.; Racicot, K.; Clark, D.S.; Austin, K.J.; Hansen, T.R.; Lucy, M.C.; Davies, C.J.; Ott, T.L. Regulation of Interferon-Stimulated Genes in Peripheral Blood Leukocytes in Pregnant and Bred, Nonpregnant Dairy Cows. J. Dairy Sci. 2007, 90, 274–280. [Google Scholar] [CrossRef]
- Pugliesi, G.; Miagawa, B.T.; Paiva, Y.N.; França, M.R.; Silva, L.A.; Binelli, M. Conceptus-Induced Changes in the Gene Expression of Blood Immune Cells and the Ultrasound-Accessed Luteal Function in Beef Cattle: How Early Can We Detect Pregnancy? Biol. Reprod. 2014, 91. [Google Scholar] [CrossRef]
- Kamat, M.M.; Vasudevan, S.; Maalouf, S.A.; Townson, D.H.; Pate, J.L.; Ott, T.L. Changes in Myeloid Lineage Cells in the Uterus and Peripheral Blood of Dairy Heifers During Early Pregnancy. Biol. Reprod. 2016, 95. [Google Scholar] [CrossRef]
- Vasudevan, S.; Kamat, M.M.; Walusimbi, S.S.; Pate, J.L.; Ott, T.L. Effects of early pregnancy on uterine lymphocytes and endometrial expression of immune-regulatory molecules in dairy heifers. Biol. Reprod. 2017, 97, 104–118. [Google Scholar] [CrossRef]
- Sanchez, J.M.; Mathew, D.J.; Behura, S.K.; Passaro, C.; Charpigny, G.; Butler, S.T.; Spencer, T.E.; Lonergan, P. Bovine endometrium responds differentially to age-matched short and long conceptusesdagger. Biol. Reprod. 2019, 101, 26–39. [Google Scholar] [CrossRef]
- Mathew, D.J.; Sánchez, J.M.; Passaro, C.; Charpigny, G.; Behura, S.K.; Spencer, T.E.; Lonergan, P. Interferon tau-dependent and independent effects of the bovine conceptus on the endometrial transcriptomedagger. Biol. Reprod. 2019, 100, 365–380. [Google Scholar] [CrossRef]
- Forde, N.; Bazer, F.W.; Spencer, T.E.; Lonergan, P. ‘Conceptualizing’ the Endometrium: Identification of Conceptus-Derived Proteins During Early Pregnancy in Cattle. Biol. Reprod. 2015, 92, 156. [Google Scholar] [CrossRef]
- Masters, R.A.; Roberts, R.M.; Lewis, G.S.; Thatcher, W.W.; Bazer, F.W.; Godkin, J.D. High molecular weight glycoproteins released by expanding, pre-attachment sheep, pig and cow blastocysts in culture. Reproduction 1982, 66, 571. [Google Scholar] [CrossRef] [Green Version]
- Burns, G.W.; Brooks, K.E.; O’Neil, E.V.; Hagen, D.E.; Behura, S.K.; Spencer, T.E. Progesterone effects on extracellular vesicles in the sheep uterus. Biol. Reprod. 2018, 98, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, I.; Xu, J.; Wang, X.; Burghardt, R.C.; Dunlap, K.A.; Bazer, F.W. Exosomes, endogenous retroviruses and toll-like receptors: Pregnancy recognition in ewes. Reproduction 2015, 149, 281. [Google Scholar] [CrossRef] [PubMed]
- Kusama, K.; Nakamura, K.; Bai, R.; Nagaoka, K.; Sakurai, T.; Imakawa, K. Intrauterine exosomes are required for bovine conceptus implantation. Biochem. Biophys. Res. Commun. 2018, 495, 1370–1375. [Google Scholar] [CrossRef] [PubMed]
- Taqi, M.O.; Saeed-Zidane, M.; Gebremedhn, S.; Salilew-Wondim, D.; Khdrawy, O.; Rings, F.; Neuhoff, C.; Hoelker, M.; Schellander, K.; Tesfaye, D. Sexual dimorphic expression and release of transcription factors in bovine embryos exposed to oxidative stress. Mol. Reprod. Dev. 2019, 86, 2005–2019. [Google Scholar] [CrossRef] [PubMed]
- Forde, N.; McGettigan, P.A.; Mehta, J.P.; O’Hara, L.; Mamo, S.; Bazer, F.W.; Spencer, T.E.; Lonergan, P. Proteomic analysis of uterine fluid during the pre-implantation period of pregnancy in cattle. Reproduction 2014, 147, 575–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexopoulos, N.I.; Vajta, G.; Maddox-Hyttel, P.; French, A.J.; Trounson, A.O. Stereomicroscopic and histological examination of bovine embryos following extended in vitro culture. Reprod. Fertil. Dev. 2005, 17, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Brandao, D.O.; Maddox-Hyttel, P.; Løvendahl, P.; Rumpf, R.; Stringfellow, D.; Callesen, H. Post hatching development: A novel system for extended in vitro culture of bovine embryos. Biol. Reprod. 2004, 71, 2048–2055. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Erdjument-Bromage, H.; Neubert, T.A. Quantitative Comparison of Proteomes Using SILAC. Curr. Protoc. Protein Sci. 2019, 95, e74. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Y.; Li, B.; Wang, H.; Guan, F.; Tan, Z.; Li, X. Screening differentially expressed proteins from co-cultured hematopoietic cells and bone marrow-derived stromal cells by quantitative proteomics (SILAC) method. Clin. Proteom. 2019, 16, 32. [Google Scholar] [CrossRef]
- Gonneaud, A.; Jones, C.; Turgeon, N.; Lévesque, D.; Asselin, C.; Boudreau, F.; Boisvert, F.M. A SILAC-Based Method for Quantitative Proteomic Analysis of Intestinal Organoids. Sci. Rep. 2016, 6, 38195. [Google Scholar] [CrossRef] [Green Version]
- Bogenhagen, D.F.; Haley, J.D. Pulse-chase SILAC-based analyses reveal selective over-synthesis and rapid turnover of mitochondrial protein components of respiratory complexes. J. Biol. Chem. 2020, 295, 2544–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beati, H.; Langlands, A.; ten Have, S.; Müller, H.A.J. SILAC-based quantitative proteomic analysis of Drosophila gastrula stage embryos mutant for fibroblast growth factor signalling. Fly 2019, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; BSherman, T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [Green Version]
- Burns, G.W.; Brooks, K.E.; Spencer, T.E. Extracellular Vesicles Originate from the Conceptus and Uterus During Early Pregnancy in Sheep. Biol. Reprod. 2016, 94. [Google Scholar] [CrossRef] [Green Version]
- Pluchino, S.; Smith, J.A. Explicating Exosomes: Reclassifying the Rising Stars of Intercellular Communication. Cell 2019, 177, 225–227. [Google Scholar] [CrossRef] [Green Version]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [Green Version]
- Bowling, H.; Bhattacharya, A.; Zhang, G.; Lebowitz, J.Z.; Alam, D.; Smith, P.T.; Kirshenbaum, K.; Neubert, T.A.; Vogel, C.; Chao, M.V.; et al. BONLAC: A combinatorial proteomic technique to measure stimulus-induced translational profiles in brain slices. Neuropharmacology 2016, 100, 76–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basisty, N.; Meyer, J.G.; Schilling, B. Protein Turnover in Aging and Longevity. Proteomics 2018, 18, e1700108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLean, J.A., 2nd; Chakrabarty, A.; Xie, S.; Bixby, J.A.; Roberts, R.M.; Green, J.A. Family of Kunitz proteins from trophoblast: Expression of the trophoblast Kunitz domain proteins (TKDP) in cattle and sheep. Mol. Reprod. Dev. 2003, 65, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Mamo, S.; Mehta, J.P.; Forde, N.; McGettigan, P.; Lonergan, P. Conceptus-endometrium crosstalk during maternal recognition of pregnancy in cattle. Biol. Reprod. 2012, 87, 6. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.N.; Potter, J.A.; Aldo, P.; Kwon, J.Y.; Pitruzzello, M.; Tong, M.; Guller, S.; Rothlin, C.V.; Mor, G.; Abrahams, V.M. Viral Infection Sensitizes Human Fetal Membranes to Bacterial Lipopolysaccharide by MERTK Inhibition and Inflammasome Activation. J. Immunol. 2017, 199, 2885–2895. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, Y.; Kiyotani, K.; Yew, P.Y.; Kato, T.; Tamura, K.; Yap, K.L.; Nielsen, S.M.; Mester, J.L.; Eng, C.; Nakamura, Y.; et al. Germline PARP4 mutations in patients with primary thyroid and breast cancers. Endocr. Relat. Cancer 2016, 23, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Prawira, A.; Munusamy, P.; Yuan, J.; Chan, C.H.T.; Koh, G.L.; Shuen, T.W.H.; Hu, J.; Yap, Y.S.; Tan, M.H.; Ang, P.; et al. Assessment of PARP4 as a candidate breast cancer susceptibility gene. Breast Cancer Res. Treat. 2019, 177, 145–153. [Google Scholar] [CrossRef]
- Cirello, V.; Colombo, C.; Pogliaghi, G.; Proverbio, M.C.; Rossi, S.; Mussani, E.; Tosi, D.; Bulfamante, G.; Bonoldi, E.; Gherardi, G.; et al. Genetic variants of PARP4 gene and PARP4P2 pseudogene in patients with multiple primary tumors including thyroid cancer. Mutat. Res. Fundam. Mol. Mech. Mutagenes. 2019, 816–818, 111672. [Google Scholar] [CrossRef]
- Oliver, F.J.; de la Rubia, G.; Rolli, V.; Ruiz-Ruiz, M.C.; de Murcia, G.; Ménissier-de Murcia, J. Importance of Poly(ADP-ribose) Polymerase and Its Cleavage in Apoptosis: Lesson from an uncleavable mutant. J. Biol. Chem. 1998, 273, 33533–33539. [Google Scholar] [CrossRef] [Green Version]
- He, T.-S.; Xie, T.; Li, J.; Yang, Y.X.; Li, C.; Wang, W.; Cao, L.; Rao, H.; Ju, C.; Xu, L.G. THO Complex Subunit 7 Homolog Negatively Regulates Cellular Antiviral Response against RNA Viruses by Targeting TBK1. Viruses 2019, 11, 158. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Xu, X.; Wu, M.; Guan, Z.; Su, X.; Chen, S.; Wang, H.; Teng, L. GPRC5A: An Emerging Biomarker in Human Cancer. BioMed Res. Int. 2018, 2018, 1823726. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jing, B.; Xu, D.; Liao, Y.; Song, H.; Sun, B.; Guo, W.; Xu, J.; Li, K.; Hu, M.; et al. PTGES/PGE2 signaling links immunosuppression and lung metastasis in Gprc5a-knockout mouse model. Oncogene 2020, 39, 3179–3194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roder, K.; Werdich, A.A.; Li, W.; Liu, M.; Kim, T.Y.; Organ-Darling, L.E.; Moshal, K.S.; Hwang, J.M.; Lu, Y.; Choi, B.-R.; et al. RING finger protein RNF207, a novel regulator of cardiac excitation. J. Biol. Chem. 2014, 289, 33730–33740. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Mabwi, H.A.; Palange, N.J.; Jia, R.; Ma, J.; Bah, F.B.; Sah, R.K.; Li, D.; Wang, D.; Bah, F.B.M.; et al. Expression Patterns and Potential Biological Roles of Dip2a. PLoS ONE 2015, 10, e0143284. [Google Scholar] [CrossRef] [PubMed]
- Fellous, A.; Earley, R.L.; Silvestre, F. The Kdm/Kmt gene families in the self-fertilizing mangrove rivulus fish, Kryptolebias marmoratus, suggest involvement of histone methylation machinery in development and reproduction. Gene 2019, 687, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Melero, R.; Hug, N.; López-Perrote, A.; Yamashita, A.; Cáceres, J.F.; Llorca, O. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat. Commun. 2016, 7, 10585. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Kwon, S.K.; Lee, S.Y.; Baek, K.H. Ubiquitin-specific peptidase 5 and ovarian tumor deubiquitinase 6A are differentially expressed in p53+/+ and p53-/-HCT116 cells. Int. J. Oncol. 2018, 52, 1705–1714. [Google Scholar]
- Yoo, H.H.; Chung, I.K. Requirement of DDX39 DEAD box RNA helicase for genome integrity and telomere protection. Aging Cell 2011, 10, 557–571. [Google Scholar] [CrossRef]
- Lingner, J.; Cooper, J.; Cech, T. Telomerase and DNA end replication: No longer a lagging strand problem? Science 1995, 269, 1533–1534. [Google Scholar] [CrossRef]
- Cerone, M.A.; Autexier, C.; Londono-Vallejo, J.A.; Bacchetti, S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene 2005, 24, 7893–7901. [Google Scholar] [CrossRef] [Green Version]
- Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 17 April 2020).




| Number of Conceptuses | Proteins | % of Total | Heavy Labelled Proteins | % Heavy Labelled Proteins | % Heavy/Total Proteins |
|---|---|---|---|---|---|
| 8 | 1576 | 14.17 | 0 | 0 | 0 |
| ≥7 | 2180 | 19.60 | 0 | 0 | 0 |
| ≥6 | 2757 | 24.79 | 0 | 0 | 0 |
| ≥5 | 3314 | 29.80 | 2 | 0.3 | 0.01 |
| ≥4 | 3997 | 35.94 | 3 | 0.45 | 0.02 |
| ≥3 | 4939 | 44.41 | 9 | 1.36 | 0.08 |
| ≥2 | 6680 | 60.06 | 51 | 7.7 | 0.46 |
| ≥1 (total) | 11,122 | 100 | 662 | 100 | 5.49 |
| GO Terms | Count | % | p Value | List Total | Pop Hits | Pop Total | Fold Enrichment |
|---|---|---|---|---|---|---|---|
| Proteasome | 31 | 2.032787 | 3.80 × 10−17 | 926 | 46 | 7550 | 5.494647 |
| Citrate cycle (TCA cycle) | 17 | 1.114754 | 6.21 × 10−8 | 926 | 30 | 7550 | 4.62023 |
| Pyruvate metabolism | 21 | 1.377049 | 1.83 × 10−9 | 926 | 38 | 7550 | 4.505797 |
| Other glycan degradation | 11 | 0.721311 | 4.21 × 10−5 | 926 | 20 | 7550 | 4.484341 |
| Propanoate metabolism | 14 | 0.918033 | 2.89 × 10−6 | 926 | 26 | 7550 | 4.390264 |
| Carbon metabolism | 55 | 3.606557 | 7.77 × 10−22 | 926 | 109 | 7550 | 4.114075 |
| Amino sugar and nucleotide sugar metabolism | 24 | 1.57377 | 1.23 × 10−9 | 926 | 48 | 7550 | 4.076674 |
| 2-Oxocarboxylic acid metabolism | 9 | 0.590164 | 6.95 × 10−4 | 926 | 18 | 7550 | 4.076674 |
| Sulfur metabolism | 5 | 0.327869 | 0.025586 | 926 | 10 | 7550 | 4.076674 |
| Glyoxylate and dicarboxylate metabolism | 12 | 0.786885 | 1.18 × 10−4 | 926 | 26 | 7550 | 3.763084 |
| Protein | Protein Name | Protein ID | B | C | D | E | F | H | I | J | Count H Labelled | Count L Labelled | Total Count |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| THO complex subunit 7 homolog | THOC7 | F1MSQ1 | H | H | L | H | 0 | 0 | H | H | 5 | 1 | 6 |
| RING finger protein 207 | RNF207 | A0JNG4 | H | H | H | 0 | H | 0 | H | 0 | 5 | 0 | 5 |
| SET domain containing 9 | SETD9 | F1MWB0 | H | H | H | L | L | H | L | 0 | 4 | 3 | 7 |
| MER proto-oncogene, tyrosine kinase | MERTK | F1N381 | H | L | H | L | L | H | L | L | 3 | 5 | 8 |
| G protein-coupled receptor class C group 5 member A | GPRC5A | F1N6N2 | H | H | 0 | L | L | H | 0 | 0 | 3 | 2 | 5 |
| DExH-box helicase 34 | DHX34 | E1BJ90 | 0 | H | H | 0 | 0 | 0 | H | 0 | 3 | 0 | 3 |
| Disco interacting protein 2 homolog A | DIP2A | F1MPW1 | 0 | 0 | 0 | H | 0 | 0 | H | H | 3 | 0 | 3 |
| Uncharacterized protein | G3X6J6 | H | 0 | 0 | H | 0 | 0 | H | 0 | 3 | 0 | 3 | |
| OTU deubiquitinase 6A | OTUD6A | G5E596 | H | 0 | H | 0 | 0 | 0 | H | 0 | 3 | 0 | 3 |
| KEGG Pathway | Count | % | p Value | List Total | Pop Hits | Pop Total | Fold Enrichment |
|---|---|---|---|---|---|---|---|
| Bile secretion | 3 | 6.818182 | 0.006783 | 15 | 68 | 7550 | 22.20588 |
| Aldosterone synthesis and secretion | 3 | 6.818182 | 0.009068 | 15 | 79 | 7550 | 19.11392 |
| Salivary secretion | 3 | 6.818182 | 0.009973 | 15 | 83 | 7550 | 18.19277 |
| Ovarian steroidogenesis | 2 | 4.545455 | 0.088901 | 15 | 50 | 7550 | 20.13333 |
| Group | Concentration (Particles/mL) | Mean Diameter (nm) | Mode Diameter (nm) |
|---|---|---|---|
| Pregnant Day 16 Heifer 1 | 1.25 × 1012 | 128.7 | 99.1 |
| Pregnant Day 16 Heifer 2 | 3.16 × 1011 | 98.1 | 95.4 |
| Pregnant Day 16 Heifer 3 | 1.04 × 1012 | 127.4 | 92.4 |
| Pregnant Day 16 Heifer 4 | 6.01 × 1011 | 111.2 | 91.6 |
| Pregnant Day 16 Heifer 5 | 1.91 × 1012 | 138.5 | 106.7 |
| Pregnant Day 16 Heifer 6 | 1.32 × 1012 | 138.8 | 106.7 |
| Pregnant Day 16 Heifer 7 | 1.63 × 1012 | 141.5 | 100.0 |
| Cyclic Day 16 Heifer 1 | 2.86 × 1011 | 115.2 | 92.1 |
| Cyclic Day 16 Heifer 2 | 1.34 × 1012 | 146.7 | 103.3 |
| Cyclic Day 16 Heifer 3 | 2.04 × 1012 | 151.1 | 108.8 |
| Cyclic Day 16 Heifer 4 | 9.01 × 1011 | 123.4 | 100.0 |
| Cyclic Day 16 Heifer 5 | 1.05 × 1012 | 162.0 | 113.3 |
| Cyclic Day 16 Heifer 6 | 1.05 × 1012 | 111.4 | 101.8 |
| KEGG Pathway | Count | % | p Value | List Total | Pop Hits | Pop Total | Fold Enrichment |
|---|---|---|---|---|---|---|---|
| Collecting duct acid secretion | 3 | 5.172414 | 7.59 × 10−3 | 38 | 27 | 7550 | 22.07602 |
| Synaptic vesicle cycle | 6 | 10.34483 | 1.22 × 10−5 | 38 | 63 | 7550 | 18.92231 |
| Endocrine and other factor-regulated calcium reabsorption | 3 | 5.172414 | 1.78 × 10−2 | 38 | 42 | 7550 | 14.19173 |
| Tight junction | 3 | 5.172414 | 0.067422 | 38 | 87 | 7550 | 6.85118 |
| Rheumatoid arthritis | 3 | 5.172414 | 0.078554 | 38 | 95 | 7550 | 6.274238 |
| Systemic lupus erythematosus | 5 | 8.62069 | 1.08 × 10−2 | 38 | 178 | 7550 | 5.581017 |
| Carbon metabolism | 3 | 5.172414 | 0.099294 | 38 | 109 | 7550 | 5.468373 |
| Phagosome | 4 | 6.896552 | 4.16 × 10−2 | 38 | 158 | 7550 | 5.02998 |
| Endocytosis | 6 | 10.34483 | 6.23 × 10−3 | 38 | 243 | 7550 | 4.905783 |
| Protein processing in endoplasmic reticulum | 4 | 6.896552 | 0.049135 | 38 | 169 | 7550 | 4.702585 |
| Biosynthesis of antibiotics | 4 | 6.896552 | 0.079032 | 38 | 206 | 7550 | 3.857946 |
| KEGG Pathway | Count | % | p Value | List Total | Pop Hits | Pop Total | Fold Enrichment |
|---|---|---|---|---|---|---|---|
| Glyoxylate and dicarboxylate metabolism | 9 | 1.978022 | 6.96 × 10−6 | 317 | 26 | 7550 | 8.244358 |
| Proteasome | 15 | 3.296703 | 2.71 × 10−9 | 317 | 46 | 7550 | 7.766424 |
| Other glycan degradation | 6 | 1.318681 | 0.001147 | 317 | 20 | 7550 | 7.14511 |
| Pentose phosphate pathway | 8 | 1.758242 | 9.06 × 10−5 | 317 | 27 | 7550 | 7.056899 |
| 2-Oxocarboxylic acid metabolism | 5 | 1.098901 | 0.005782 | 317 | 18 | 7550 | 6.615843 |
| Ribosome | 36 | 7.912088 | 4.09 × 10−19 | 317 | 134 | 7550 | 6.398606 |
| Biosynthesis of amino acids | 19 | 4.175824 | 3.97 × 10−10 | 317 | 71 | 7550 | 6.373573 |
| Citrate cycle (TCA cycle) | 8 | 1.758242 | 1.86 × 10−4 | 317 | 30 | 7550 | 6.351209 |
| Cysteine and methionine metabolism | 10 | 2.197802 | 1.96 × 10−5 | 317 | 38 | 7550 | 6.267641 |
| Glycosaminoglycan degradation | 6 | 1.318681 | 0.002246 | 317 | 23 | 7550 | 6.213139 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malo Estepa, I.; Tinning, H.; Rosas Vasconcelos, E.J.; Fernandez-Fuertes, B.; Sánchez, J.M.; Burns, G.W.; Spencer, T.E.; Lonergan, P.; Forde, N. Protein Synthesis by Day 16 Bovine Conceptuses during the Time of Maternal Recognition of Pregnancy. Int. J. Mol. Sci. 2020, 21, 2870. https://doi.org/10.3390/ijms21082870
Malo Estepa I, Tinning H, Rosas Vasconcelos EJ, Fernandez-Fuertes B, Sánchez JM, Burns GW, Spencer TE, Lonergan P, Forde N. Protein Synthesis by Day 16 Bovine Conceptuses during the Time of Maternal Recognition of Pregnancy. International Journal of Molecular Sciences. 2020; 21(8):2870. https://doi.org/10.3390/ijms21082870
Chicago/Turabian StyleMalo Estepa, Irene, Haidee Tinning, Elton Jóse Rosas Vasconcelos, Beatriz Fernandez-Fuertes, José María Sánchez, Gregory W. Burns, Thomas E. Spencer, Pat Lonergan, and Niamh Forde. 2020. "Protein Synthesis by Day 16 Bovine Conceptuses during the Time of Maternal Recognition of Pregnancy" International Journal of Molecular Sciences 21, no. 8: 2870. https://doi.org/10.3390/ijms21082870





