Comparison of Target Recognition by TRAF1 and TRAF2
Abstract
:1. Introduction
2. Results
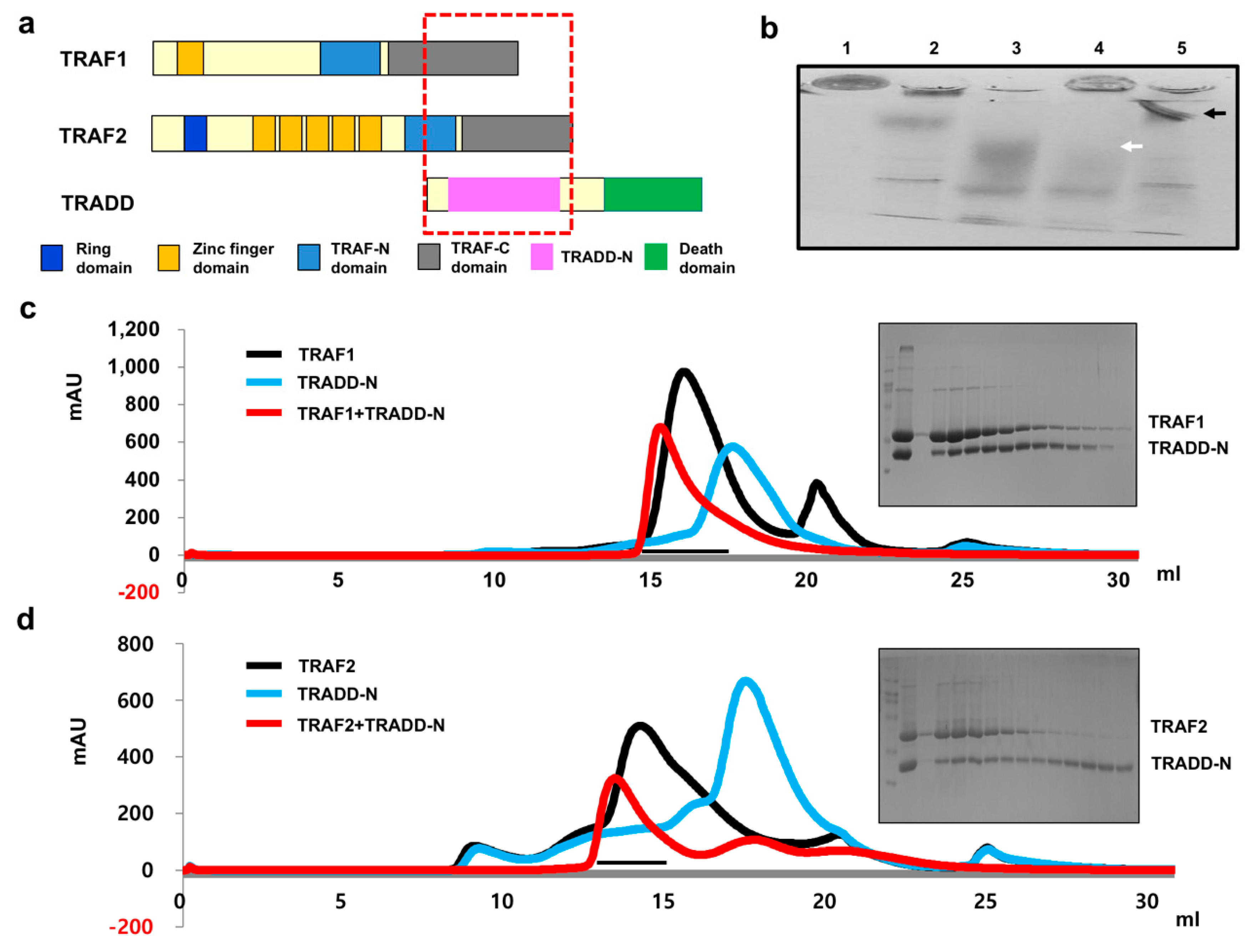
2.1. Both TRAF1 and TRAF2 Interact with TRADD via Their TRAF Domain In Vitro
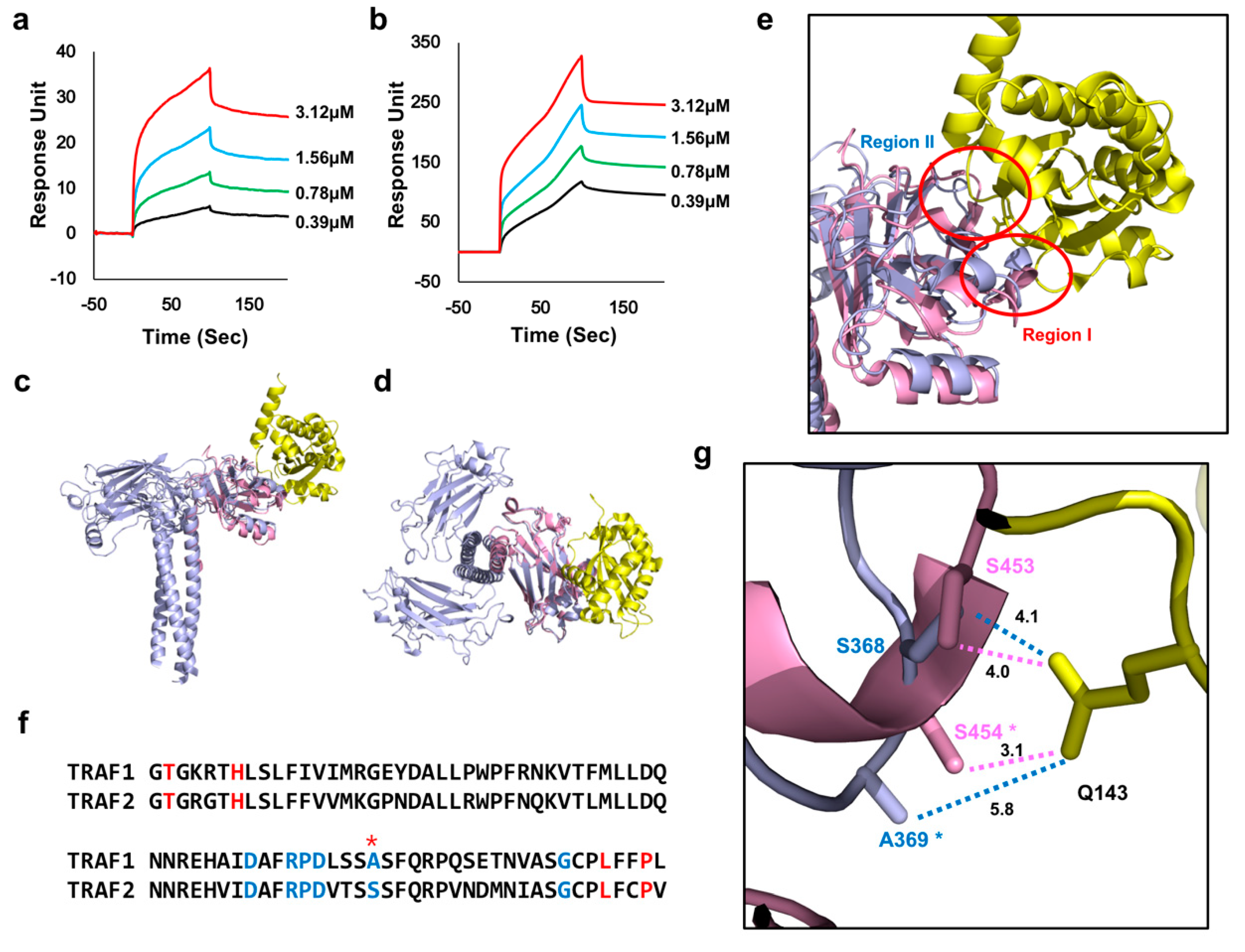
2.2. TRADD Interacts with TRAF2 with Higher Affinity than TRAF1
2.3. TANK and Caspase-2 Interact more Tightly with TRAF1 than TRAF2
2.4. F347 on TRAF1 Positively Effects the Affinity of TANK and Caspase-2
3. Discussion
4. Materials and Methods
4.1. Peptide Preparation
4.2. Visualization of Structure Models
4.3. Sequence Alignment
4.4. Protein Expression and Purification
4.5. Native-PAGE
4.6. Complex Formation Assay by Gel-filtration Chromatography
4.7. Surface Plasmon Resonance (SPR)
4.8. Isothermal Titration Calorimetry (ITC)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rothe, M.; Sarma, V.; Dixit, V.M.; Goeddel, D.V. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 1995, 269, 1424–1427. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Arron, J.R.; Lamothe, B.; Cirilli, M.; Kobayashi, T.; Shevde, N.K.; Segal, D.; Dzivenu, O.K.; Vologodskaia, M.; Yim, M.; et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 2002, 418, 443–447. [Google Scholar] [CrossRef]
- Park, H.H. Structure of TRAF Family: Current Understanding of Receptor Recognition. Front. Immunol. 2018, 9, 1999. [Google Scholar] [CrossRef] [Green Version]
- Zapata, J.M.; Pawlowski, K.; Haas, E.; Ware, C.F.; Godzik, A.; Reed, J.C. TEFs: A diverse family of proteins containing TRAF domains. J. Biol. Chem. 2001, 276, 24242–24252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uren, A.G.; Vaux, D.L. TRAF proteins and meprins share a conserved domain. Trends Biochem. Sci. 1996, 21, 244–245. [Google Scholar] [CrossRef]
- Alvarez, S.E.; Harikumar, K.B.; Hait, N.C.; Allegood, J.; Strub, G.M.; Kim, E.Y.; Maceyka, M.; Jiang, H.; Luo, C.; Kordula, T.; et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 2010, 465, 1084–1088. [Google Scholar] [CrossRef] [Green Version]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu Rev. Biochem 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Greenfeld, H.; Takasaki, K.; Walsh, M.J.; Ersing, I.; Bernhardt, K.; Ma, Y.; Fu, B.; Ashbaugh, C.W.; Cabo, J.; Mollo, S.B.; et al. TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation. PLoS Pathog. 2015, 11, e1004890. [Google Scholar] [CrossRef] [Green Version]
- Inoue, J.; Ishida, T.; Tsukamoto, N.; Kobayashi, N.; Naito, A.; Azuma, S.; Yamamoto, T. Tumor necrosis factor receptor-associated factor (TRAF) family: Adapter proteins that mediate cytokine signaling. Exp. Cell Res. 2000, 254, 14–24. [Google Scholar] [CrossRef]
- Chung, J.Y.; Park, Y.C.; Ye, H.; Wu, H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002, 115, 679–688. [Google Scholar]
- Bradley, J.R.; Pober, J.S. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 2001, 20, 6482–6491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, G.A. The multifaceted roles of TRAFs in the regulation of B-cell function. Nat. Rev. Immunol. 2004, 4, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Baud, V.; Liu, Z.G.; Bennett, B.; Suzuki, N.; Xia, Y.; Karin, M. Signaling by proinflammatory cytokines: Oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999, 13, 1297–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.; Baltimore, D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes Dev. 1996, 10, 963–973. [Google Scholar] [CrossRef] [Green Version]
- Natoli, G.; Costanzo, A.; Ianni, A.; Templeton, D.J.; Woodgett, J.R.; Balsano, C.; Levrero, M. Activation of SAPK/JNK by TNF receptor 1 through a noncytotoxic TRAF2-dependent pathway. Science 1997, 275, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, J.L.; Baltimore, D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. Embo J. 1999, 18, 6694–6704. [Google Scholar] [CrossRef]
- Robeson, A.C.; Lindblom, K.R.; Wojton, J.; Kornbluth, S.; Matsuura, K. Dimer-specific immunoprecipitation of active caspase-2 identifies TRAF proteins as novel activators. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Bhat, E.A.; Kim, C.M.; Kim, S.; Park, H.H. In Vitro Inhibitory Mechanism Effect of TRAIP on the Function of TRAF2 Revealed by Characterization of Interaction Domains. Int. J. Mol. Sci. 2018, 19, 2457. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.; Shu, H.B.; Pan, M.G.; Goeddel, D.V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996, 84, 299–308. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.; Xiong, J.; Goeddel, D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kB activation. Cell 1995, 81, 495–504. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.C.; Ye, H.; Hsia, C.; Segal, D.; Rich, R.L.; Liou, H.-L.; Myszka, D.G.; Wu, H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell 2000, 101, 777–787. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Choi, Y. TRAF1 and its biological functions. Adv. Exp. Med. Biol. 2007, 597, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Zapata, J.M.; Reed, J.C. TRAF1: Lord without a RING. Sci STKE 2002, 2002, pe27. [Google Scholar] [CrossRef]
- Tsitsikov, E.N.; Laouini, D.; Dunn, I.F.; Sannikova, T.Y.; Davidson, L.; Alt, F.W.; Geha, R.S. TRAF1 is a negative regulator of TNF signaling. enhanced TNF signaling in TRAF1-deficient mice. Immunity 2001, 15, 647–657. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.Y.; Li, Z.Z.; Jiang, D.S.; Wang, L.; Zhang, Y.; Chen, K.; Zhang, X.F.; Liu, Y.; Fan, G.C.; Chen, Y.; et al. TRAF1 is a critical regulator of cerebral ischaemia-reperfusion injury and neuronal death. Nat. Commun 2013, 4, 2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, M.; Wang, P.X.; Wang, A.B.; Zhang, X.J.; Zhang, Y.; Zhang, P.; Mei, F.H.; Chen, M.H.; Li, H. Targeting hepatic TRAF1-ASK1 signaling to improve inflammation, insulin resistance, and hepatic steatosis. J. hepatology 2016, 64, 1365–1377. [Google Scholar] [CrossRef]
- Kim, C.M.; Choi, J.Y.; Bhat, E.A.; Jeong, J.H.; Son, Y.J.; Kim, S.; Park, H.H. Crystal structure of TRAF1 TRAF domain and its implications in the TRAF1-mediated intracellular signaling pathway. Sci. Rep. 2016, 6, 25526. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.C.; Burkitt, V.; Villa, A.R.; Tong, L.; Wu, H. Structural basis for self-association and receptor recognition of human TRAF2. Nature 1999, 398, 533–538. [Google Scholar] [CrossRef]
- Zhang, P.; Reichardt, A.; Liang, H.H.; Aliyari, R.; Cheng, D.; Wang, Y.Y.; Xu, F.; Cheng, G.H.; Liu, Y.F. Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5. Sci. Signal. 2012, 5, ra81. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.M.; Son, Y.J.; Kim, S.; Kim, S.Y.; Park, H.H. Molecular basis for unique specificity of human TRAF4 for platelets GPIbbeta and GPVI. Proc. Natl. Acad. Sci. USA 2017, 114, 11422–11427. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Cho, Y.J.; Park, H.H. Structure of the TRAF4 TRAF domain with a coiled-coil domain and its implications for the TRAF4 signalling pathway. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Tsao, D.H.; McDonagh, T.; Telliez, J.B.; Hsu, S.; Malakian, K.; Xu, G.Y.; Lin, L.L. Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway. Mol. Cell 2000, 5, 1051–1057. [Google Scholar] [CrossRef]
- Kim, C.M.; Jeong, J.H.; Son, Y.J.; Choi, J.H.; Kim, S.; Park, H.H. Molecular basis for TANK recognition by TRAF1 revealed by the crystal structure of TRAF1/TANK complex. FEBS Lett. 2017, 591, 810–821. [Google Scholar] [CrossRef] [Green Version]
- Foight, G.W.; Keating, A.E. Comparison of the peptide binding preferences of three closely related TRAF paralogs: TRAF2, TRAF3, and TRAF5. Protein Sci. 2016, 25, 1273–1289. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.M.; Park, H.H. Comparison of Target Recognition by TRAF1 and TRAF2. Int. J. Mol. Sci. 2020, 21, 2895. https://doi.org/10.3390/ijms21082895
Kim CM, Park HH. Comparison of Target Recognition by TRAF1 and TRAF2. International Journal of Molecular Sciences. 2020; 21(8):2895. https://doi.org/10.3390/ijms21082895
Chicago/Turabian StyleKim, Chang Min, and Hyun Ho Park. 2020. "Comparison of Target Recognition by TRAF1 and TRAF2" International Journal of Molecular Sciences 21, no. 8: 2895. https://doi.org/10.3390/ijms21082895





