Interaction of Cationic Carbosilane Dendrimers and Their siRNA Complexes with MCF-7 Cells
Abstract
:1. Introduction
2. Results
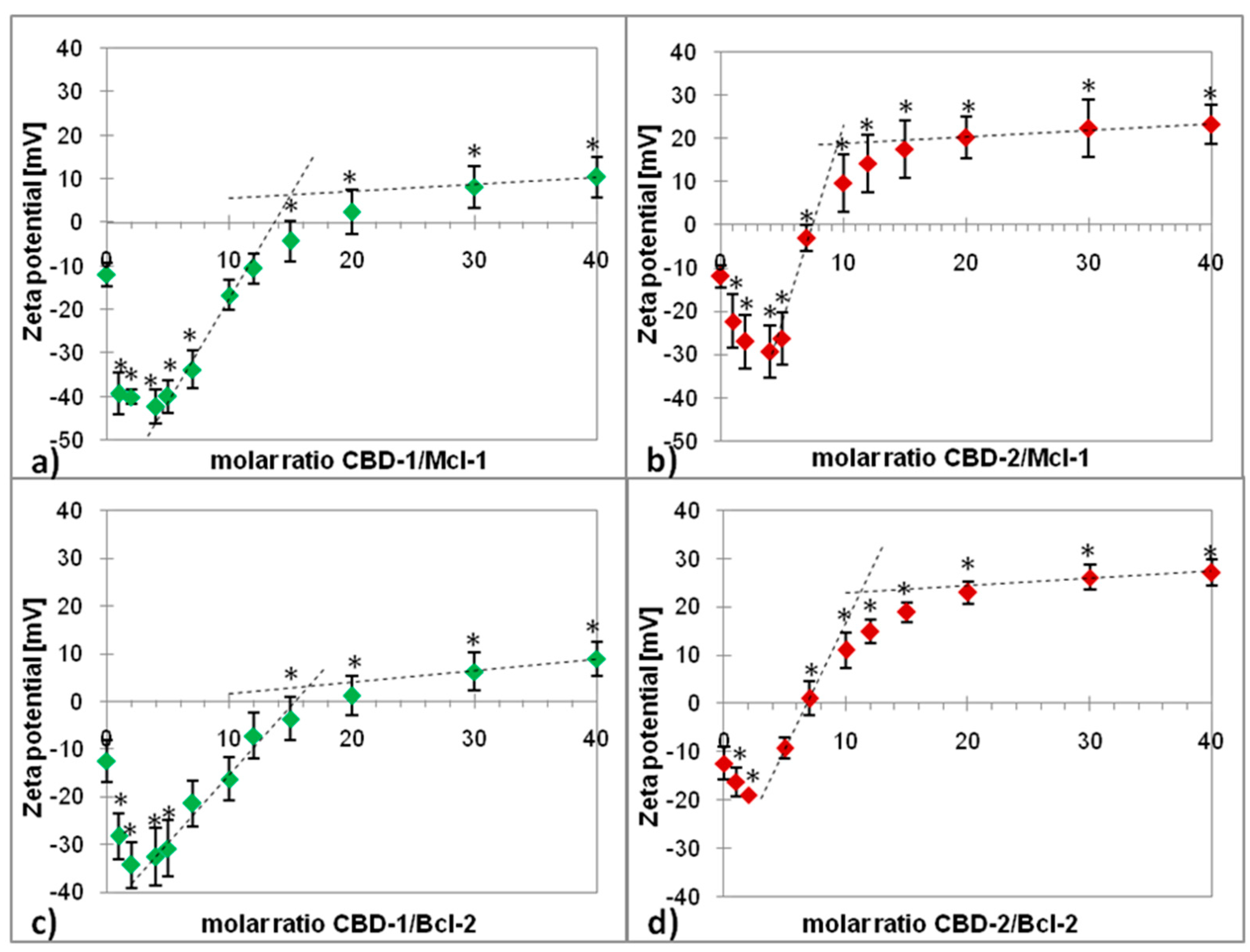
2.1. Zeta Potential
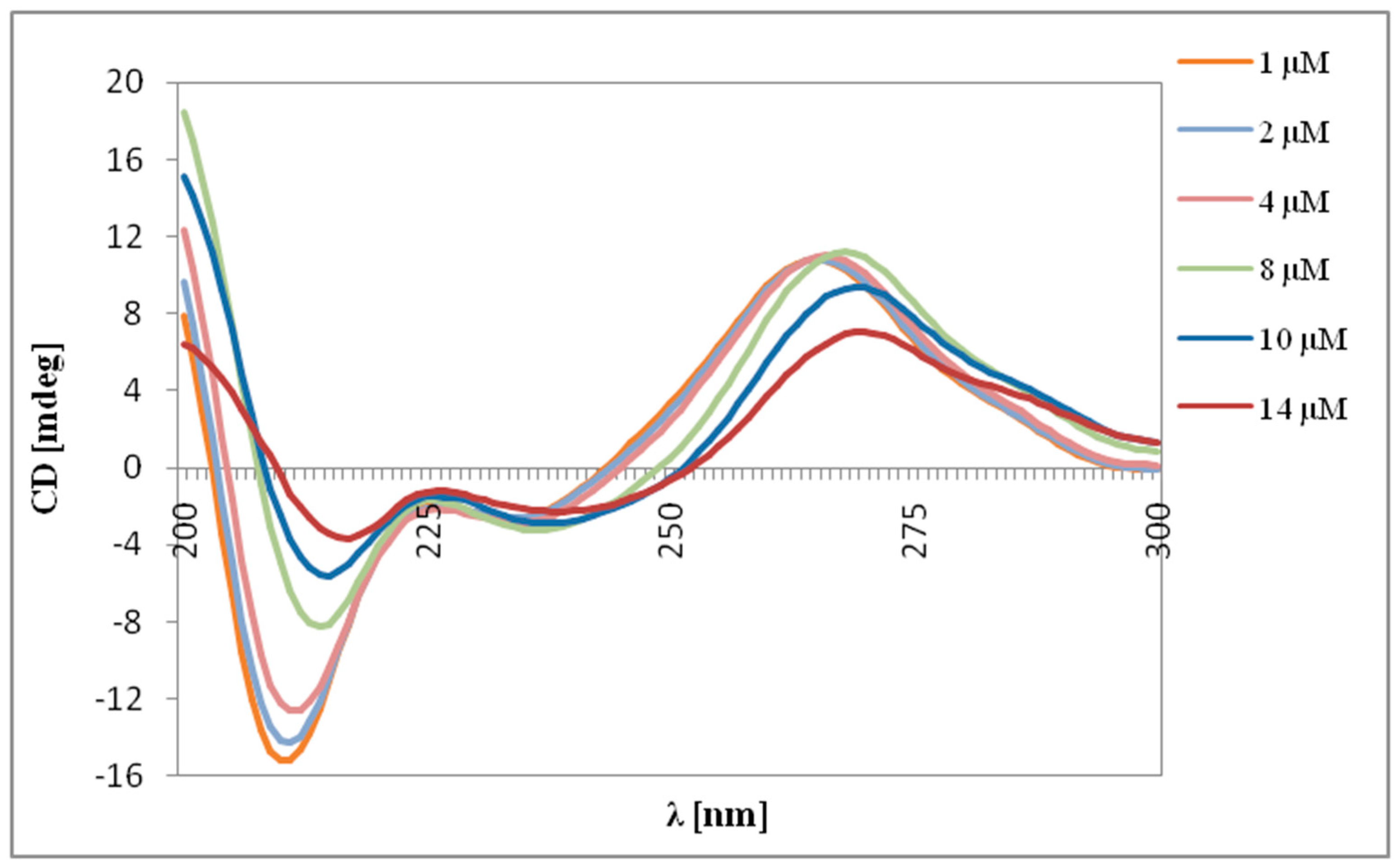
2.2. Circular Dichroism
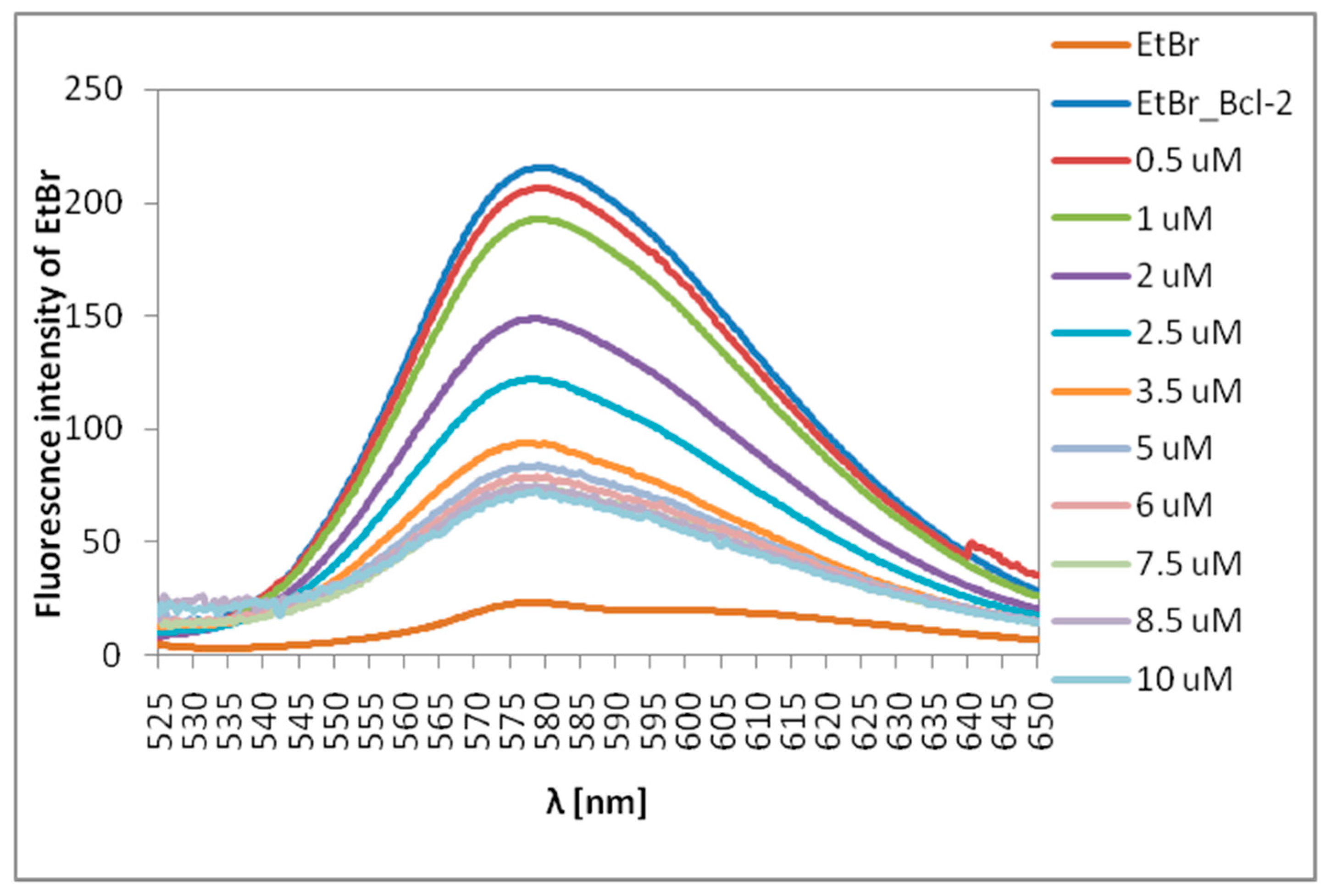
2.3. Ethidium Bromide (EtBr) Intercalation Assay
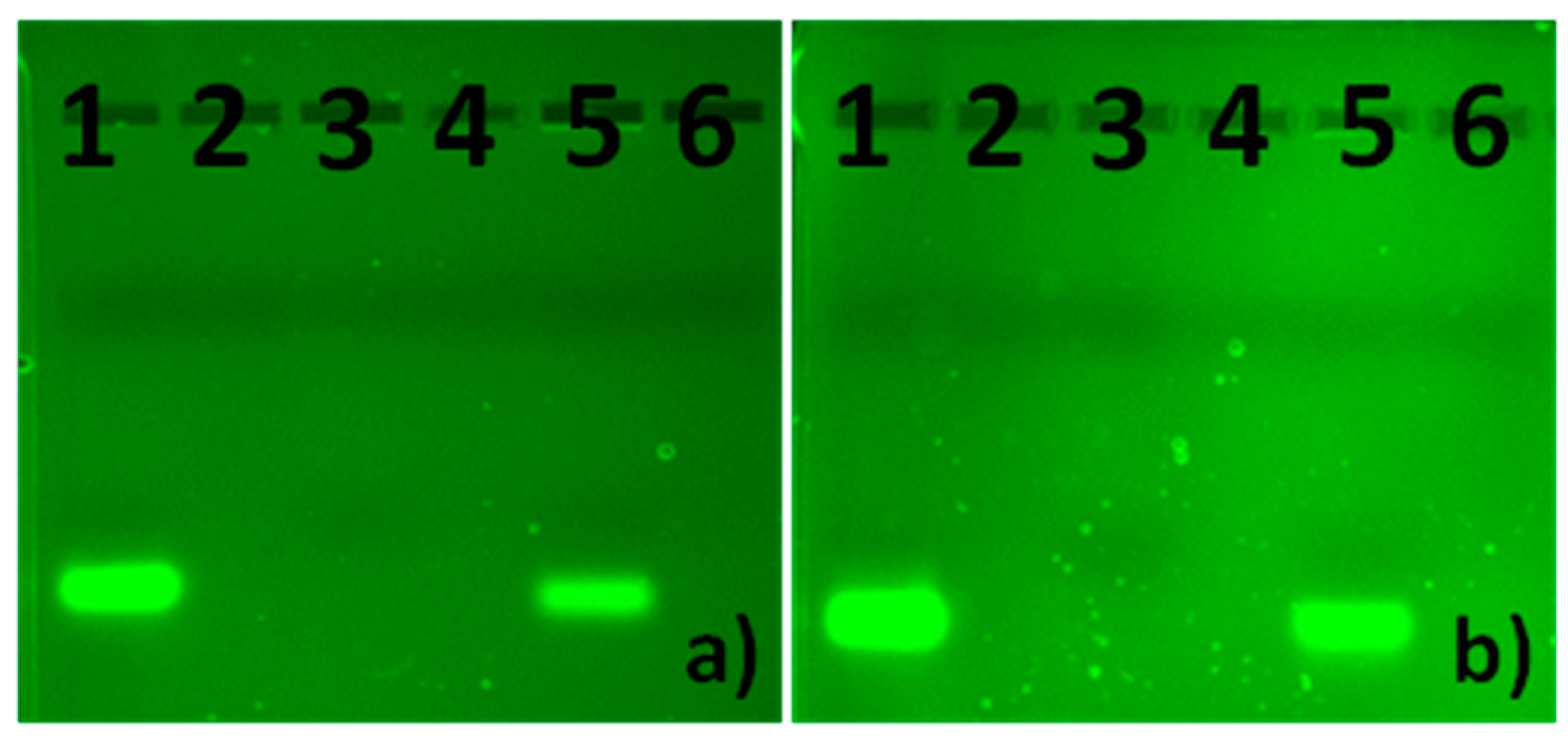
2.4. Gel Electrophoresis
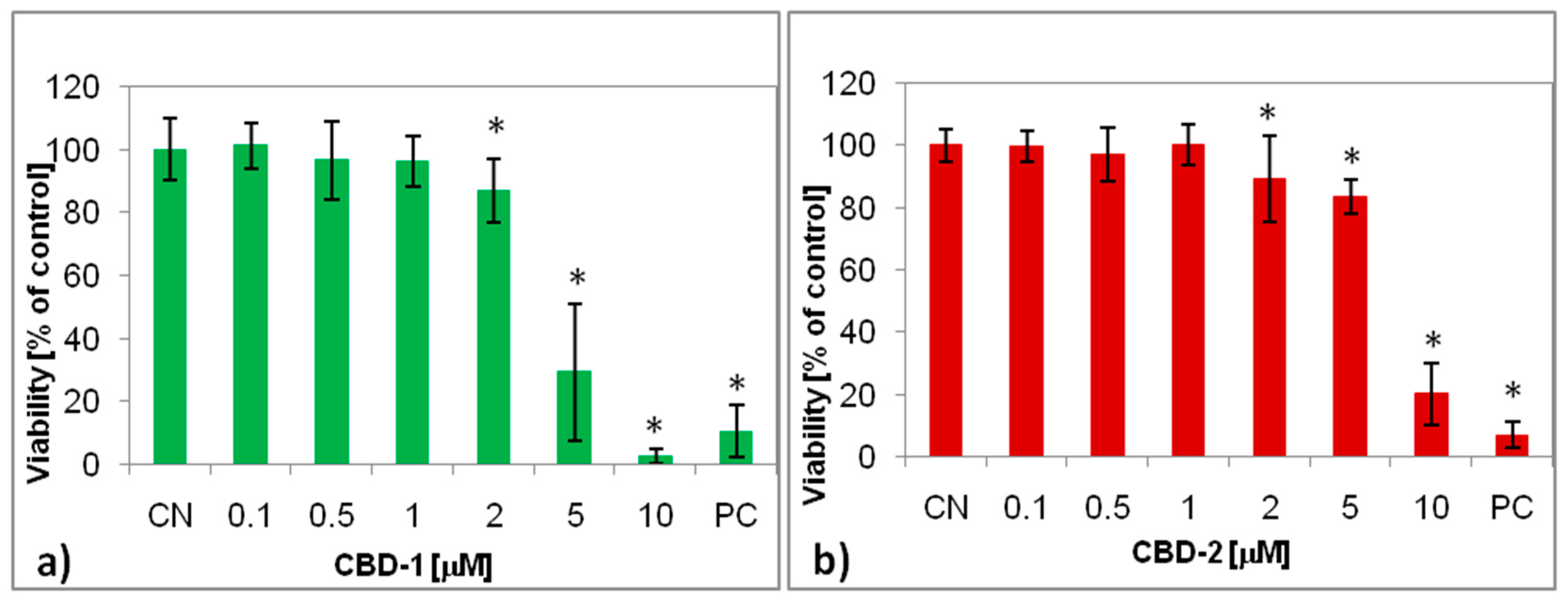
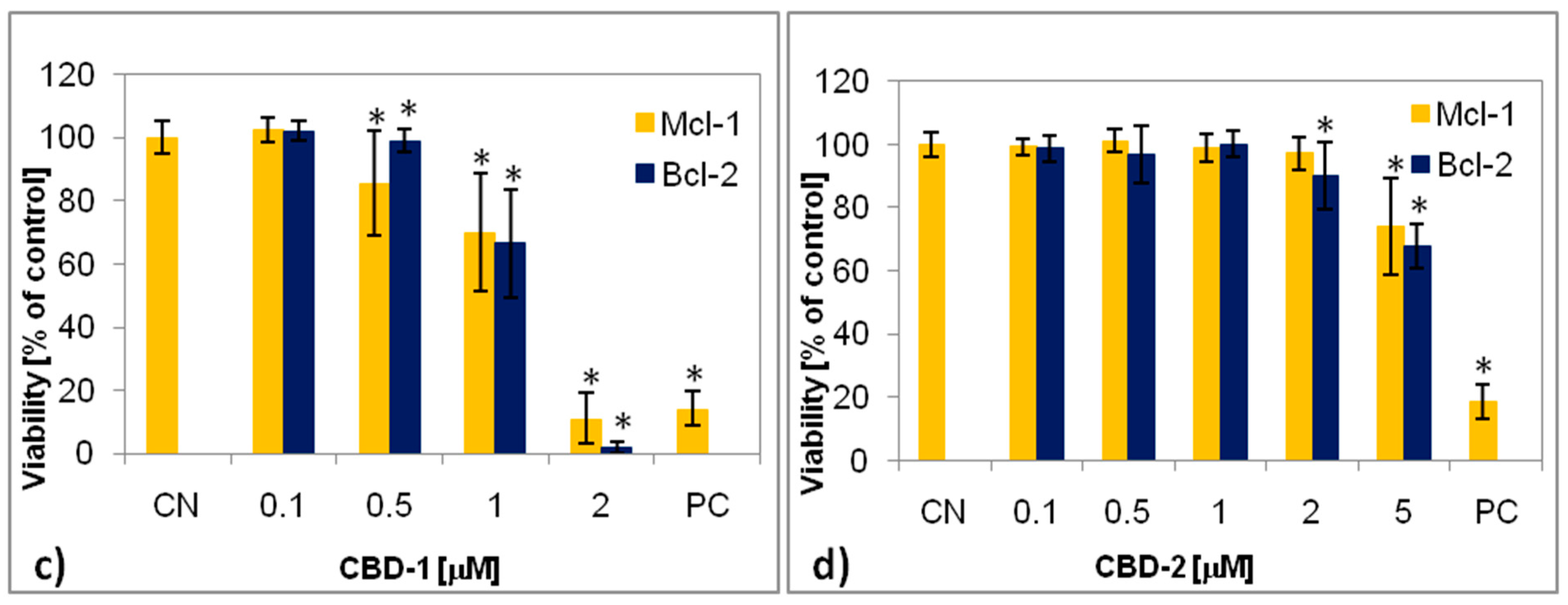
2.5. XTT Assay
2.6. Cellular Uptake
3. Discussion
4. Materials and Methods
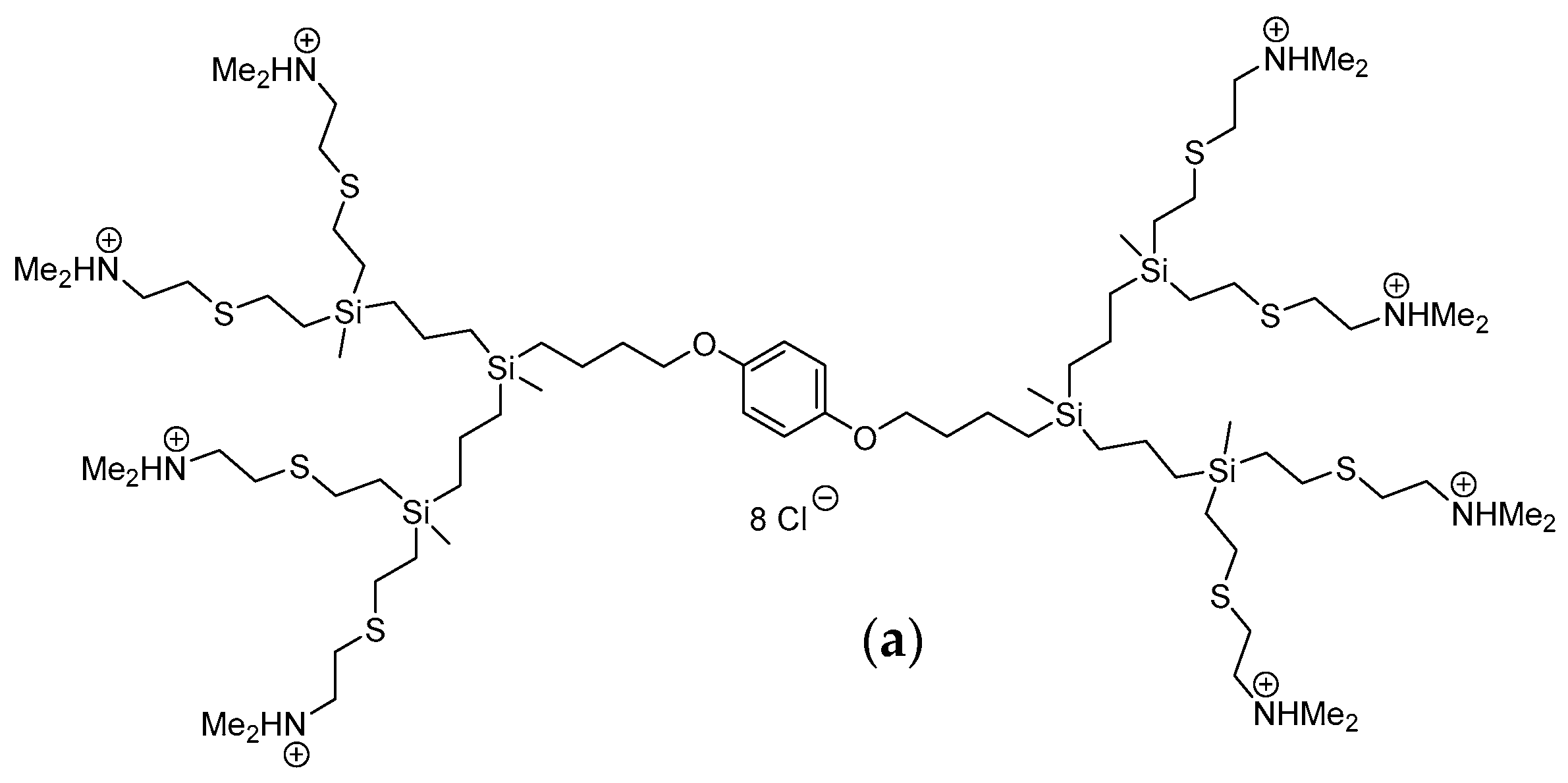
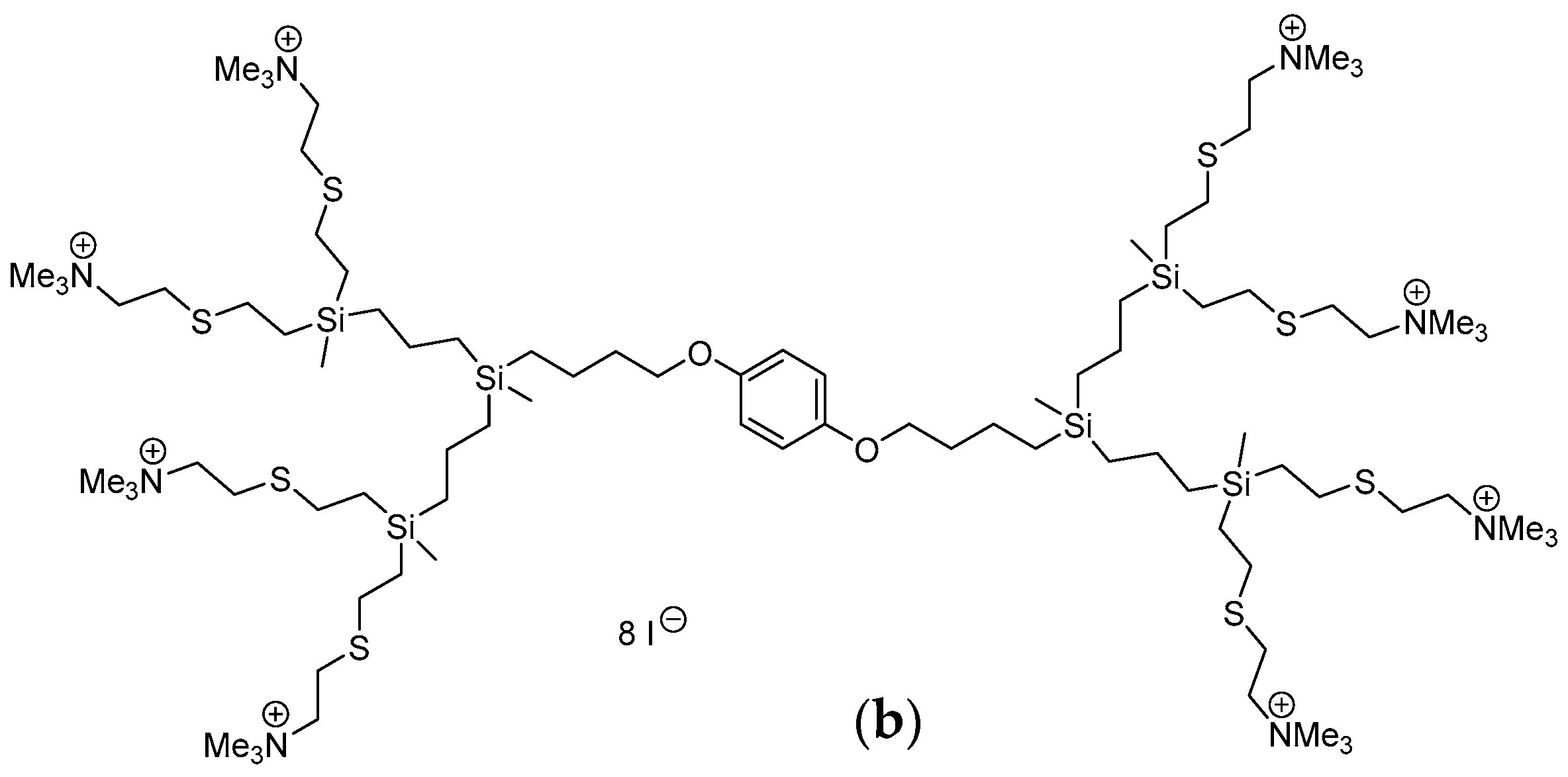
4.1. Carbosilane Dendrimers
4.2. siRNA
| Mcl-1 | |
| Sense Mcl-1s | 5′-GGACUUUUAUACCUGUUAUdTdT 3′ |
| Antisense Mcl-1a | 5′-AUAACAGGUAUAAAAGUCCdTdT 3′ |
| Bcl-2 | |
| Sense E1s | 5′-G CUG CAC CUG ACG CCC UUCdTdT 3′ |
| Antisense E1a | 5′-GAA GGG CGU CAG GUG CAG CdTdT 3′ |
4.3. Other Reagents
4.4. Dendrimer/siRNA Complexes Formation
4.5. Zeta Potential
4.6. Circular Dichroism
4.7. Ethidium Bromide (EtBr) Intercalation Assay
4.8. Gel Electrophoresis
4.9. MCF-7 Cell Line
4.10. XTT Assay
4.11. Cellular Uptake
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Vankayala, R.; Hwang, K.C. Near-infrared-light-activatable nanomaterial-mediated phototheranostic nanomedicines: An emerging paradigm for cancer treatment. Adv. Mater. 2018, 30, 1706320. [Google Scholar] [CrossRef] [PubMed]
- Michlewska, S.; Kubczak, M.; Maroto-Diaz, M.; Sanz del Olmo, N.; Ortega, P.; Shcharbin, D.; Gomez-Ramirez, R.; de la Mata, F.J.; Ionov, M.; Bryszewska, M. Synthesis and characterization of FITC labelled ruthenium dendrimer as a prospective anticancer drug. Biomolecules 2019, 9, 411. [Google Scholar] [CrossRef] [Green Version]
- Ola, M.S.; Nawaz, M.; Ahsan, H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol. Cell. Biochem. 2011, 351, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Burlacu, A. Regulation of apoptosis by Bcl-2 family proteins. J. Cell. Mol. Med. 2003, 7, 249–257. [Google Scholar] [CrossRef]
- Ionov, M.; Lazniewska, J.; Dzmitruk, V.; Halets, I.; Loznikova, S.; Novopashina, D.; Apartsin, E.; Krasheninina, O.; Venyaminova, A.; Milowska, K.; et al. Anticancer siRNA cocktails as a novel tool to treat cancer cells. Part (A). Mechanisms of interaction. Int. J. Pharm. 2015, 485, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Ambesajir, A.; Kaushik, A.; Kaushik, J.J.; Petros, S.T. RNA interference: A futuristic tool and its therapeutic applications. Saudi J. Biol. Sci. 2012, 19, 395–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angart, P.; Vocelle, D.; Chan, C.; Walton, S.P. Design of siRNA therapeutics from the molecular scale. Pharmaceuticals 2013, 6, 440–468. [Google Scholar] [CrossRef] [Green Version]
- Dzmitruk, V.; Szulc, A.; Shcharbin, D.; Janaszewska, A.; Shcharbina, N.; Lazniewska, J.; Novopashina, D.; Buyanova, M.; Ionov, M.; Klajnert-Maculewicz, B.; et al. Anticancer siRNA cocktails as a novel tool to treat cancer cells. Part (B). Efficiency of pharmacological action. Int. J. Pharm. 2015, 485, 288–294. [Google Scholar] [CrossRef]
- Chang, P.K.C.; Prestidge, C.A.; Bremmell, K.E. Interfacial analysis of siRNA complexes with poly-ethylenimine (PEI) or PAMAM dendrimers in gene delivery. Colloids Surf. B Biointerfaces 2017, 158, 370–378. [Google Scholar] [CrossRef]
- Tokatlian, T.; Segura, T. siRNA applications in nanomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 305–315. [Google Scholar] [CrossRef]
- Song, E.W.; Zhu, P.C.; Lee, S.K.; Chowdhury, D.; Kussman, S.; Dykxhoorn, D.M.; Feng, Y.; Palliser, D.; Weiner, D.B.; Shankar, P.; et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005, 23, 709–717. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, J.; Hafdi, N.; Behr, J.P.; Erbacher, P.; Peng, L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem. Commun. 2006, 22, 2362–2364. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Uppuluri, S.; Swanson, D.R.; Li, J. Dendrimers as reactive modules for the synthesis of new structure-controlled, higher-complexity megamers. Pure Appl. Chem. 2000, 72, 2343–2358. [Google Scholar] [CrossRef] [Green Version]
- Tomalia, D.A. Architecturally driven properties based on the dendritic state. High Perform. Polym. 2001, 13, S1–S10. [Google Scholar] [CrossRef]
- Sękowski, S.; Miłowska, K.; Gabryelak, T. Dendrymery w naukach biomedycznych i nanotechnologii. Postep. Hig. Med. Dosw. 2008, 62, 725–733. [Google Scholar]
- Wu, J.; Huang, W.; He, Z. Dendrimers as carriers for siRNA delivery and gene silencing: A review. Sci. World J. 2013, 2013, 630654. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lu, Z.; Wientjes, M.G.; Au, J.L.S. Delivery of siRNA therapeutics: Barriers and carriers. AAPS J. 2010, 12, 492–503. [Google Scholar] [CrossRef]
- Solassol, J.; Crozet, C.; Perrier, V.; Leclaire, J.; Beranger, F.; Caminade, A.M.; Meunier, B.; Dormont, D.; Majoral, J.P.; Lehmann, S. Cationic phosphorus-containing dendrimers reduce prion replication both in cell culture and in mice infected with scrapie. J. Gen. Virol. 2004, 85, 1791–1799. [Google Scholar] [CrossRef]
- Caminade, A.M.; Majoral, J.P. Water-soluble phosphorus-containing dendrimers. Prog. Polym. Sci. 2005, 30, 491–505. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Pedziwiatr-Werbicka, E.; Gorzkiewicz, M.; Horodecka, K.; Abashkin, V.; Klajnert-Maculewicz, B.; Pena-Gonzalez, C.E.; Sanchez-Nieves, J.; Gomez, R.; de la Mata, F.J.; Bryszewska, M. Silver nanoparticles surface-modified with carbosilane dendrons as carriers of anticancer siRNA. Int. J. Mol. Sci. 2020, 21, 4647. [Google Scholar] [CrossRef]
- Kaneda, Y. Gene therapy: A battle against biological barriers. Curr. Mol. Med. 2001, 1, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Hickerson, R.P.; Vlassov, A.V.; Wang, Q.; Leake, D.; Ilves, H.; Gonzalez-Gonzalez, E.; Contag, C.H.; Johnston, B.H.; Kaspar, R.L. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides 2008, 18, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Giacca, M.; Zacchigna, S. Virus-mediated gene delivery for human gene therapy. J. Control. Release 2012, 161, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Huang, M.; Guo, W.W.; Huang, Q.; Zhang, L.Z.; Jiang, G. Nano-based delivery of RNAi in cancer therapy. Mol. Cancer 2017, 16, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardikis, K.; Fessas, D.; Signorelli, M.; Dimas, K.; Tsimplouli, C.; Ionov, M.; Demetzos, C. A new chimeric drug delivery nano system (chi-aDDnS) composed of PAMAM G 3. 5 dendrimer and liposomes as doxorubicin’s carrier, in vitro pharmacological studies. J. Nanosci. Nanotechnol. 2011, 11, 3764–3772. [Google Scholar] [CrossRef] [PubMed]
- Shcharbin, D.; Janaszewska, A.; Klajnert-Maculewicz, B.; Ziemba, B.; Dzmitruk, V.; Halets, I.; Loznikova, S.; Shcharbina, N.; Milowska, K.; Ionov, M.; et al. How to study dendrimers and dendriplexes III. Biodistribution: Pharmacokinetics and toxicity in vivo. J. Control. Release 2014, 181, 40–52. [Google Scholar] [CrossRef]
- Białkowska, K.; Komorowski, P.; Bryszewska, M.; Miłowska, K. Spheroids as a type of three-dimensional cell cultures—Examples of methods of preparation and the most important application. Int. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef]
- Weber, N.; Ortega, P.; Clemente, M.I.; Shcharbin, D.; Bryszewska, M.; de la Mata, F.J.; Gomez, R.; Munoz-Fernandez, M.A. Characterization of carbosilane dendrimers as effective carriers of siRNA to HIV-infected lymphocytes. J. Control. Release 2008, 132, 55–64. [Google Scholar] [CrossRef]
- Posadas, I.; Lopez-Hernandez, B.; Clemente, M.I.; Jimenez, J.L.; Ortega, P.; de la Mata, J.; Gomez, R.; Munoz-Fernandez, M.A.; Cena, V. Highly efficient transfection of rat cortical neurons using carbosilane dendrimers unveils a neuroprotective role for HIF-1 alpha in early chemical hypoxia-mediated neurotoxicity. Pharm. Res. 2009, 26, 1181–1191. [Google Scholar] [CrossRef]
- Krasheninina, O.A.; Apartsin, E.K.; Fuentes, E.; Szulc, A.; Ionov, M.; Venyaminova, A.G.; Shcharbin, D.; de la Mata, F.J.; Bryszewska, M.; Gomez, R. Complexes of pro-apoptotic siRNAs and carbosilane dendrimers: Formation and effect on cancer cells. Pharmaceutics 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albertazzi, L.; Mickler, F.M.; Pavan, G.M.; Salomone, F.; Bardi, G.; Panniello, M.; Amir, E.; Kang, T.; Killops, K.L.; Brauchle, C.; et al. Enhanced bioactivity of internally functionalized cationic dendrimers with PEG cores. Biomacromolecules 2012, 13, 4089–4097. [Google Scholar] [CrossRef] [Green Version]
- Aisina, R.; Mukhametova, L.; Ivanova, E. Influence cationic and anionic PAMAM dendrimers of low generation on selected hemostatic parameters in vitro. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 109, 110605. [Google Scholar] [CrossRef]
- Wrobel, D.; Kolanowska, K.; Gajek, A.; Gomez-Ramirez, R.; de la Mata, J.; Pedziwiatr-Werbicka, E.; Klajnert, B.; Waczulikova, I.; Bryszewska, M. Interaction of cationic carbosilane dendrimers and their complexes with siRNA with erythrocytes and red blood cell ghosts. Biochim. Biophys. Acta 2014, 1838, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michlewska, S.; Ionov, M.; Maroto-Diaz, M.; Szwed, A.; Ihnatsyeu-Kachan, A.; Loznikova, S.; Shcharbin, D.; Maly, M.; Ramirez, R.G.; de la Mata, F.J.; et al. Ruthenium dendrimers as carriers for anticancer siRNA. J. Inorg. Biochem. 2018, 181, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Reveles, J.; Sedaghat-Herati, R.; Gilley, D.R.; Schaeffer, A.M.; Ghosh, K.C.; Greene, T.D.; Gann, H.E.; Dowler, W.A.; Kramer, S.; Dean, J.M.; et al. mPEG-PAMAM-G4 nucleic acid nanocomplexes: Enhanced stability, RNase protection, and activity of splice switching oligomer and Poly I:C RNA. Biomacromolecules 2013, 14, 4108–4115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jevprasesphant, R.; Penny, J.; Jalal, R.; Attwood, D.; McKeown, N.B.; D’Emanuele, A. The influence of surface modification on the cytotoxicity of PAMAM dendrimers. Int. J. Pharm. 2003, 252, 263–266. [Google Scholar] [CrossRef]
- Peña-González, C.E.; Pedziwiatr-Werbicka, E.; Shcharbin, D.; Guerrero-Beltran, C.; Abashkin, V.; Loznikova, S.; Jimenez, J.L.; Munoz-Fernandez, M.A.; Bryszewska, M.; Gomez, R.; et al. Gold nanoparticles stabilized by cationic carbosilane dendrons: Synthesis and biological properties. Dalton Trans. 2017, 46, 8736–8745. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Cruz, T.; Alcarraz-Vizan, G.; de la Mata, F.J.; de Pablo, S.; Ortega, P.; Duarte, Y.; Bravo-Moraga, F.; Gonazlez-Nilo, F.D.; Novials, A.; Gomez, R. Cationic carbosilane dendritic systems as promising anti-amyloid agents in type 2 diabetes. Chem. Eur. J. 2020, 26, 7609–7621. [Google Scholar] [CrossRef]
- Sze, A.; Erickson, D.; Ren, L.; Li, D. Zeta-potential measurement using the Smoluchowski equation and the slope of the current-time relationship in electroosmotic flow. J. Colloid Interface Sci. 2003, 261, 402–410. [Google Scholar] [CrossRef]
- Olmsted, J.; Kearns, D.R. Mechanism of ethidium-bromide fluorescence enhancement on binding to nucleic-acids. Biochemistry 1977, 16, 3647–3654. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Fink, B.E.; Brunette, S.R.; Tse, W.C.; Hedrick, M.P. A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001, 123, 5878–5891. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.; Calderón, M.; Schulz, A.; Andreou, I.; Weber, M.; Haag, R. Dendritic polyglycerols with oligoamine shells show low toxicity and high siRNA transfection efficiency in vitro. Bioconj. Chem. 2010, 21, 1744–1752. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Białkowska, K.; Miłowska, K.; Michlewska, S.; Sokołowska, P.; Komorowski, P.; Lozano-Cruz, T.; Gomez-Ramirez, R.; de la Mata, F.J.; Bryszewska, M. Interaction of Cationic Carbosilane Dendrimers and Their siRNA Complexes with MCF-7 Cells. Int. J. Mol. Sci. 2021, 22, 7097. https://doi.org/10.3390/ijms22137097
Białkowska K, Miłowska K, Michlewska S, Sokołowska P, Komorowski P, Lozano-Cruz T, Gomez-Ramirez R, de la Mata FJ, Bryszewska M. Interaction of Cationic Carbosilane Dendrimers and Their siRNA Complexes with MCF-7 Cells. International Journal of Molecular Sciences. 2021; 22(13):7097. https://doi.org/10.3390/ijms22137097
Chicago/Turabian StyleBiałkowska, Kamila, Katarzyna Miłowska, Sylwia Michlewska, Paulina Sokołowska, Piotr Komorowski, Tania Lozano-Cruz, Rafael Gomez-Ramirez, Francisco Javier de la Mata, and Maria Bryszewska. 2021. "Interaction of Cationic Carbosilane Dendrimers and Their siRNA Complexes with MCF-7 Cells" International Journal of Molecular Sciences 22, no. 13: 7097. https://doi.org/10.3390/ijms22137097





