Structure, Distribution, Chemical Composition, and Gene Expression Pattern of Glandular Trichomes on the Leaves of Rhus potaninii Maxim
Abstract
:1. Introduction
2. Results
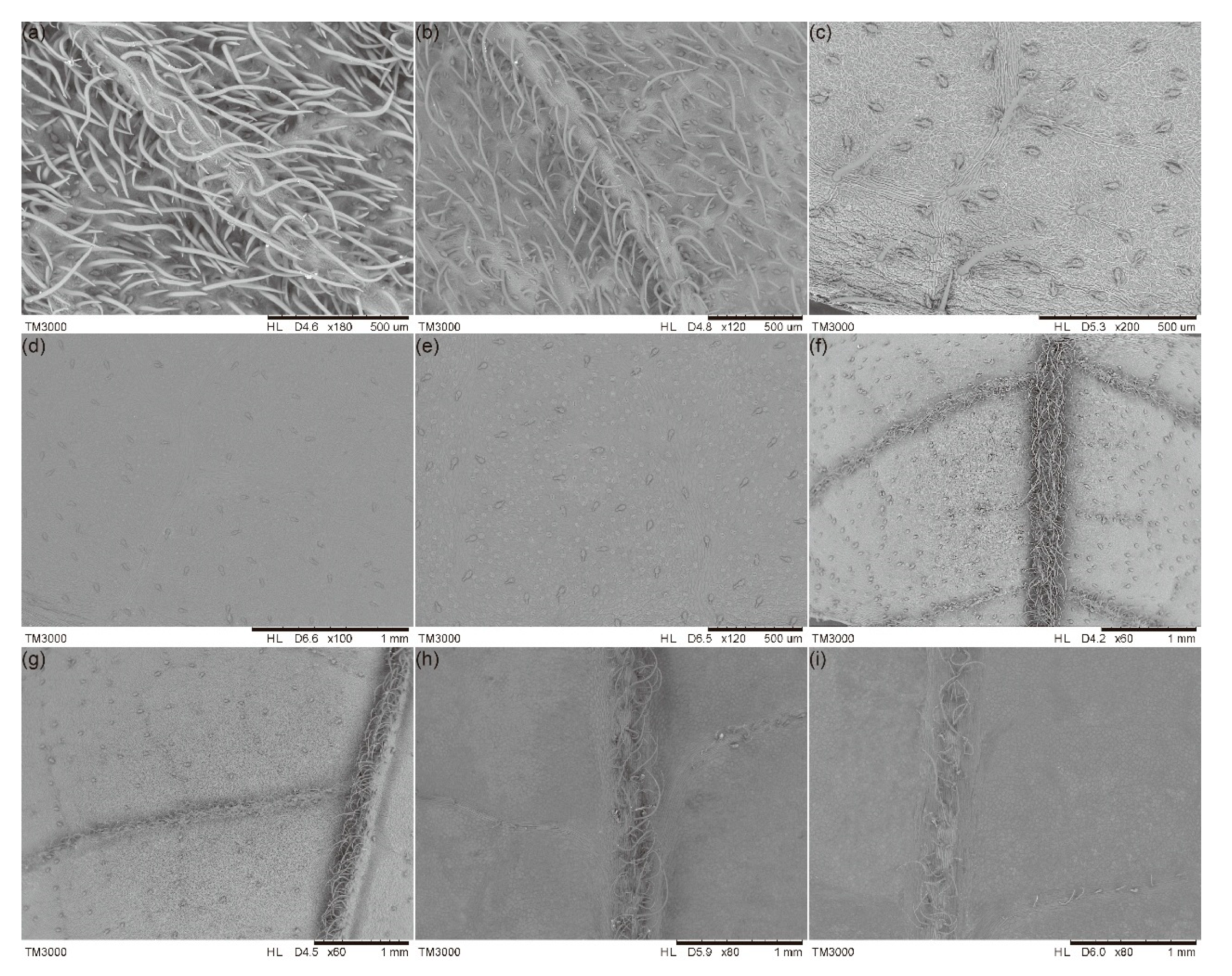
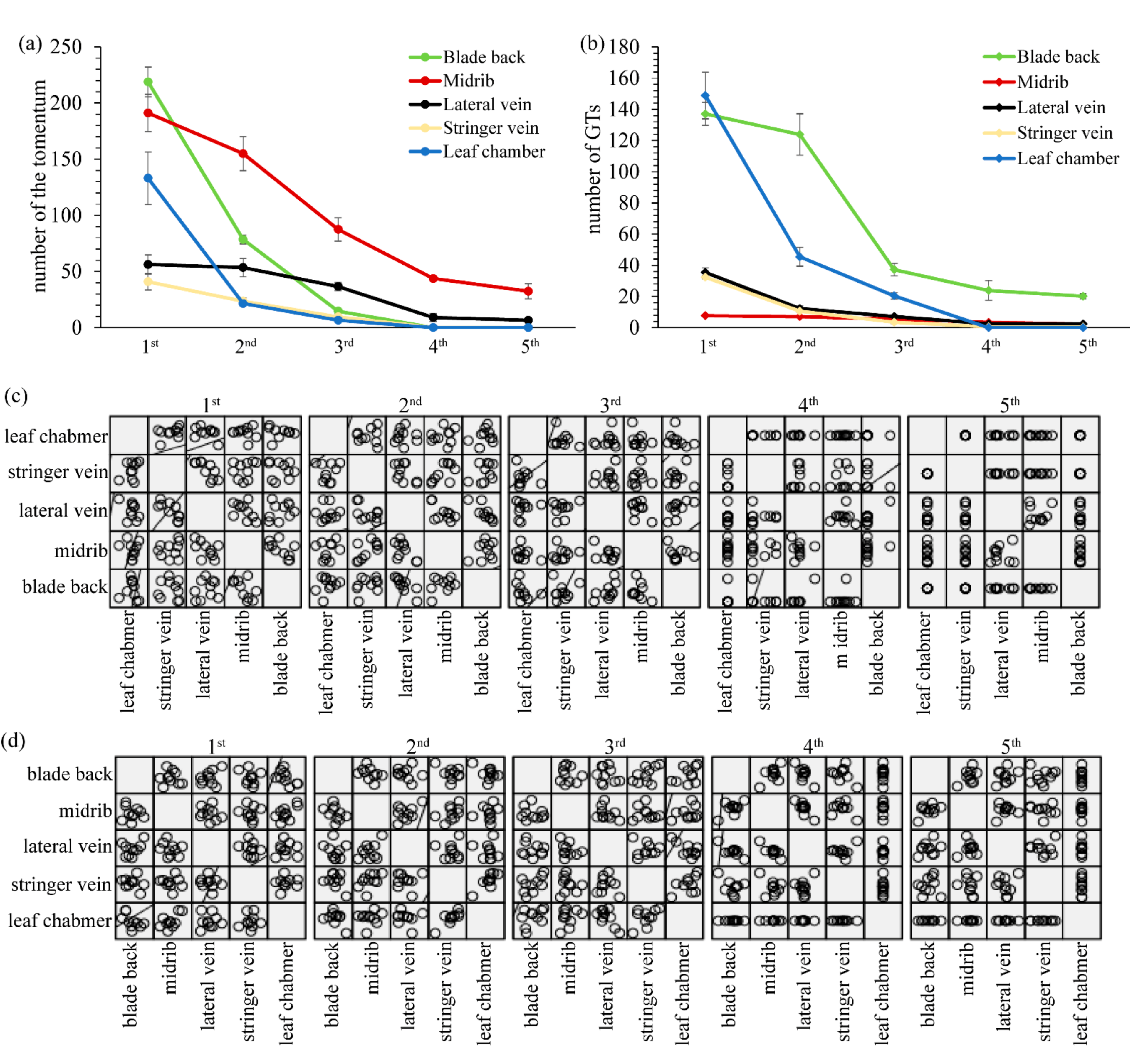
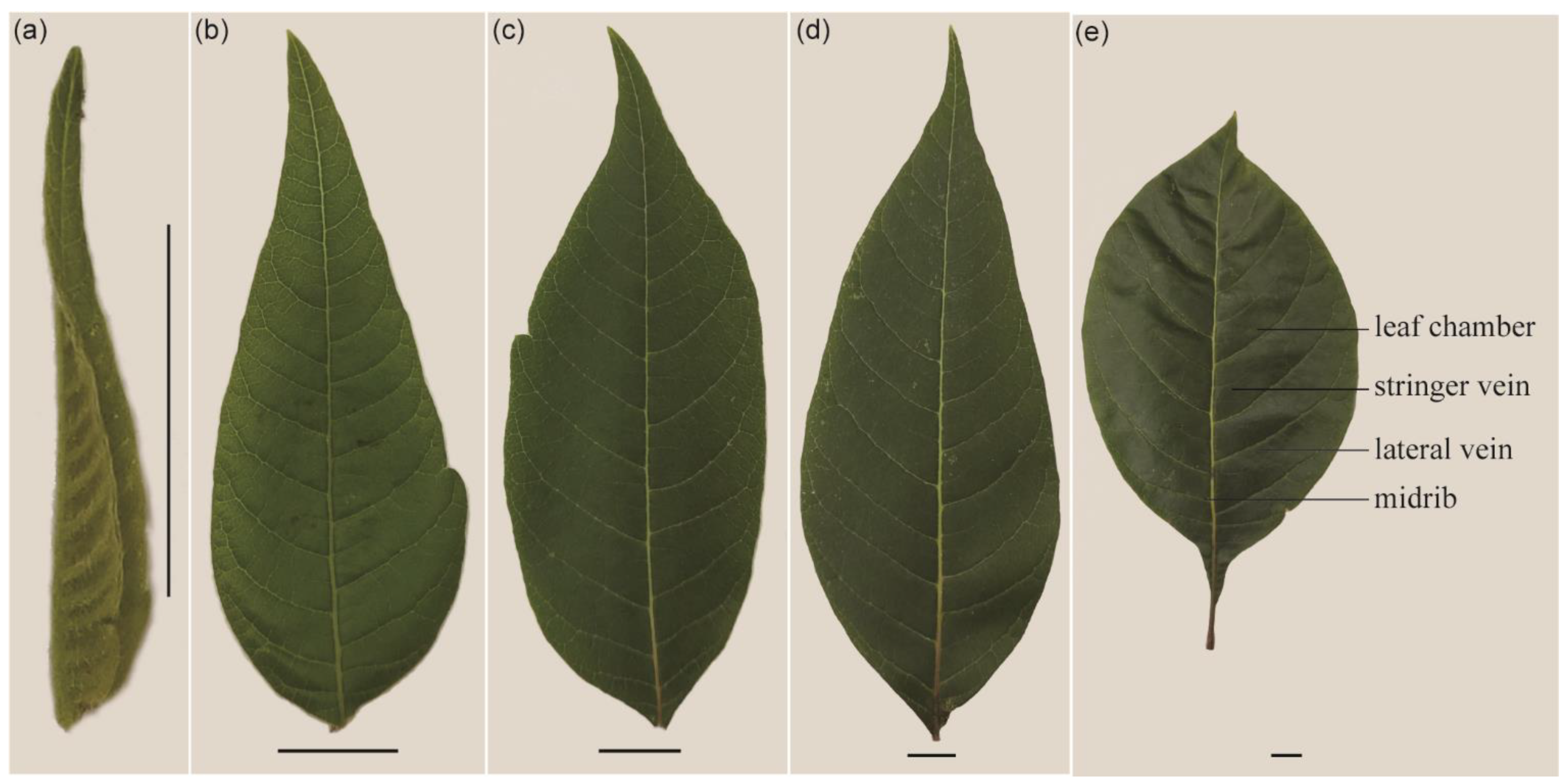
2.1. Morphological Characteristics and the Distribution of GTs
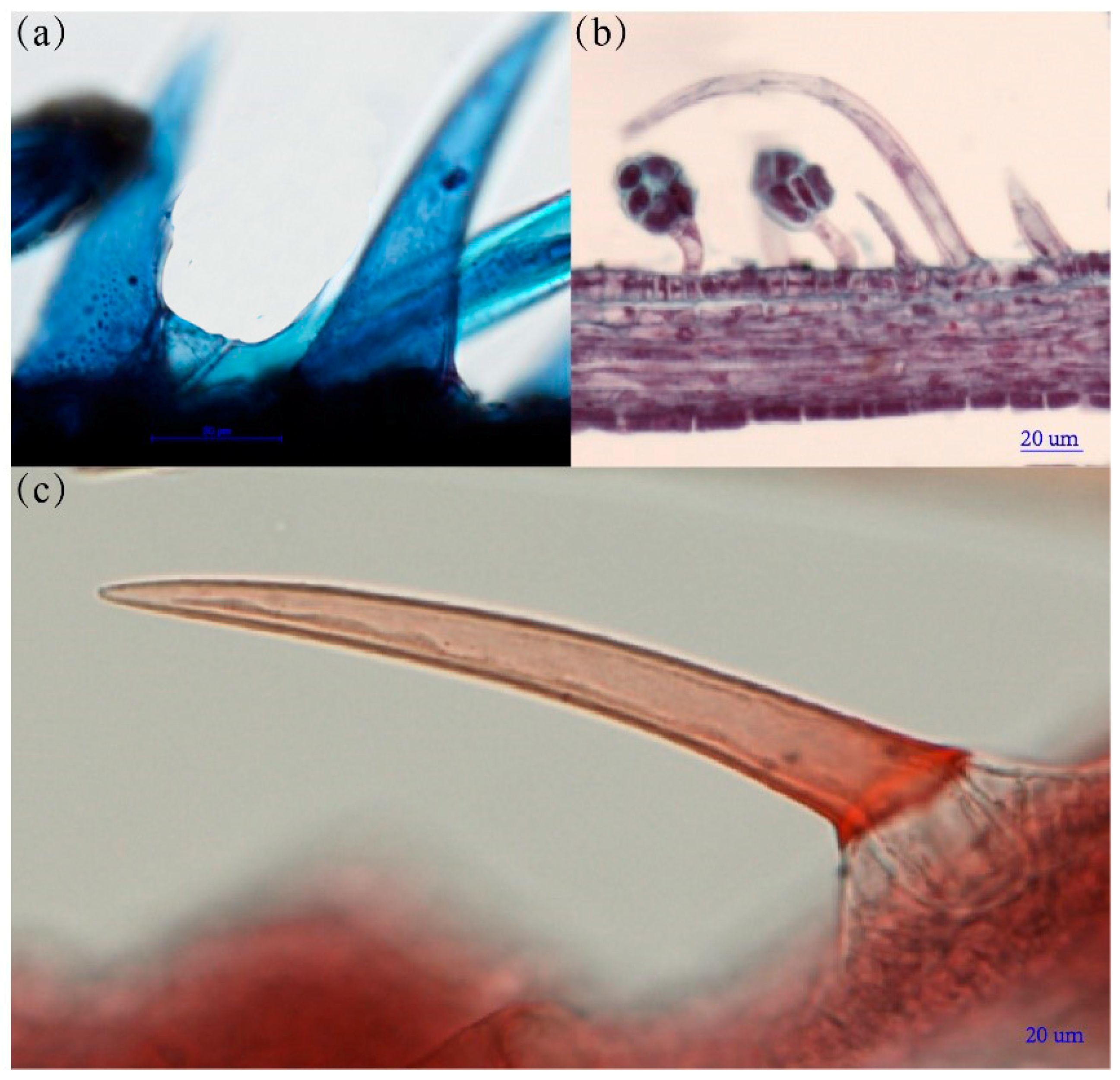
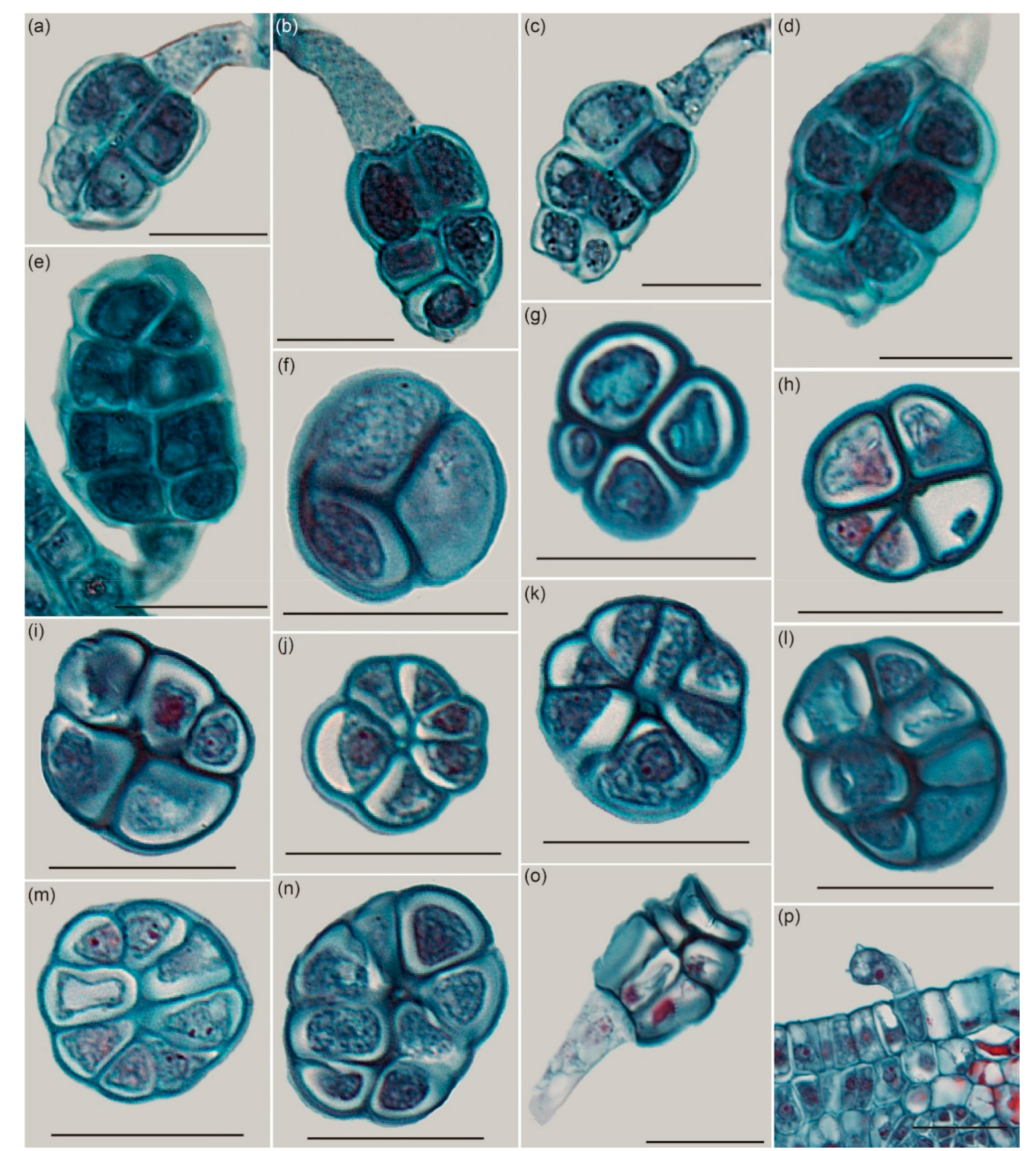
2.2. The Histological Structure of GTs
2.3. Component Analysis of GTs
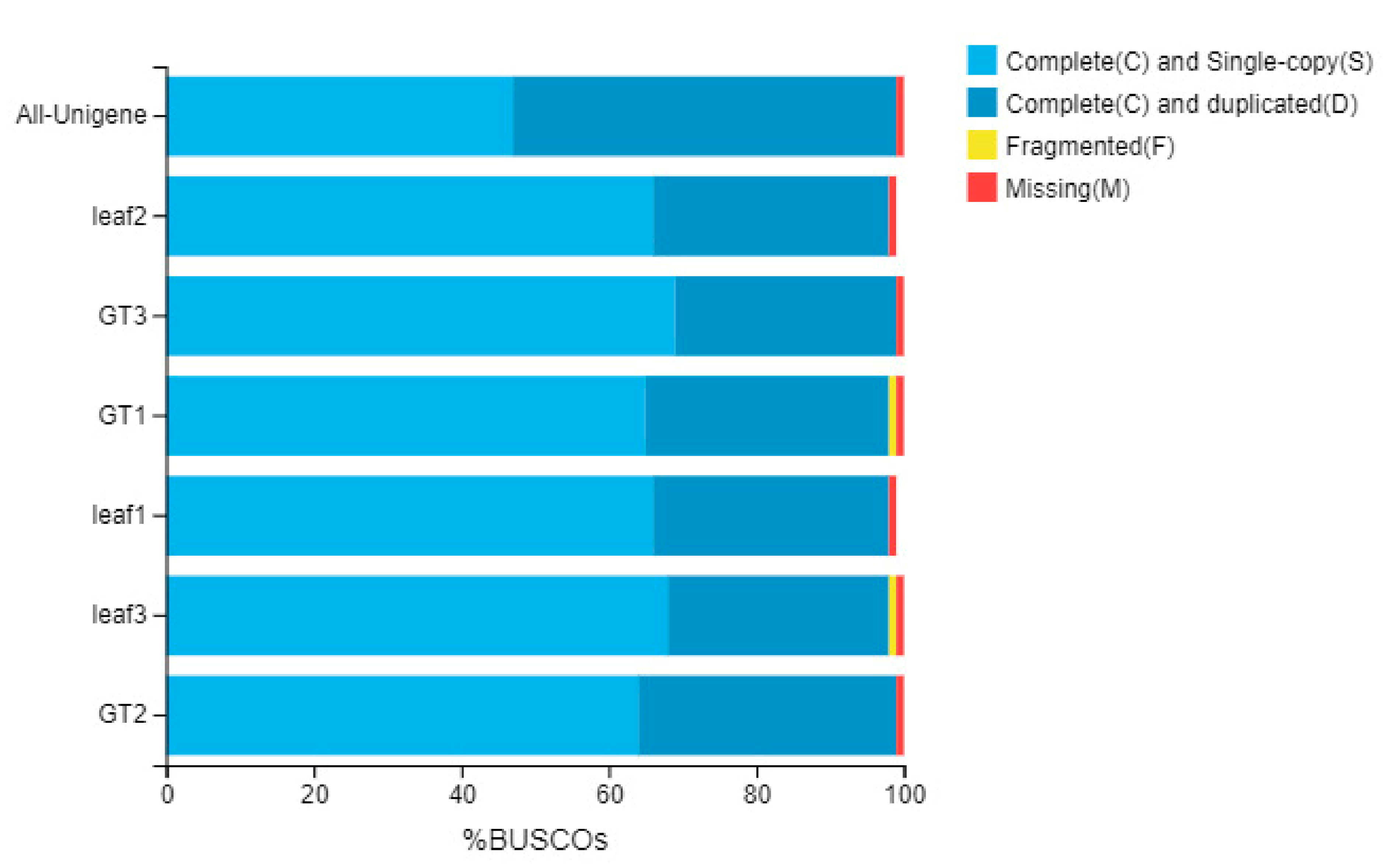
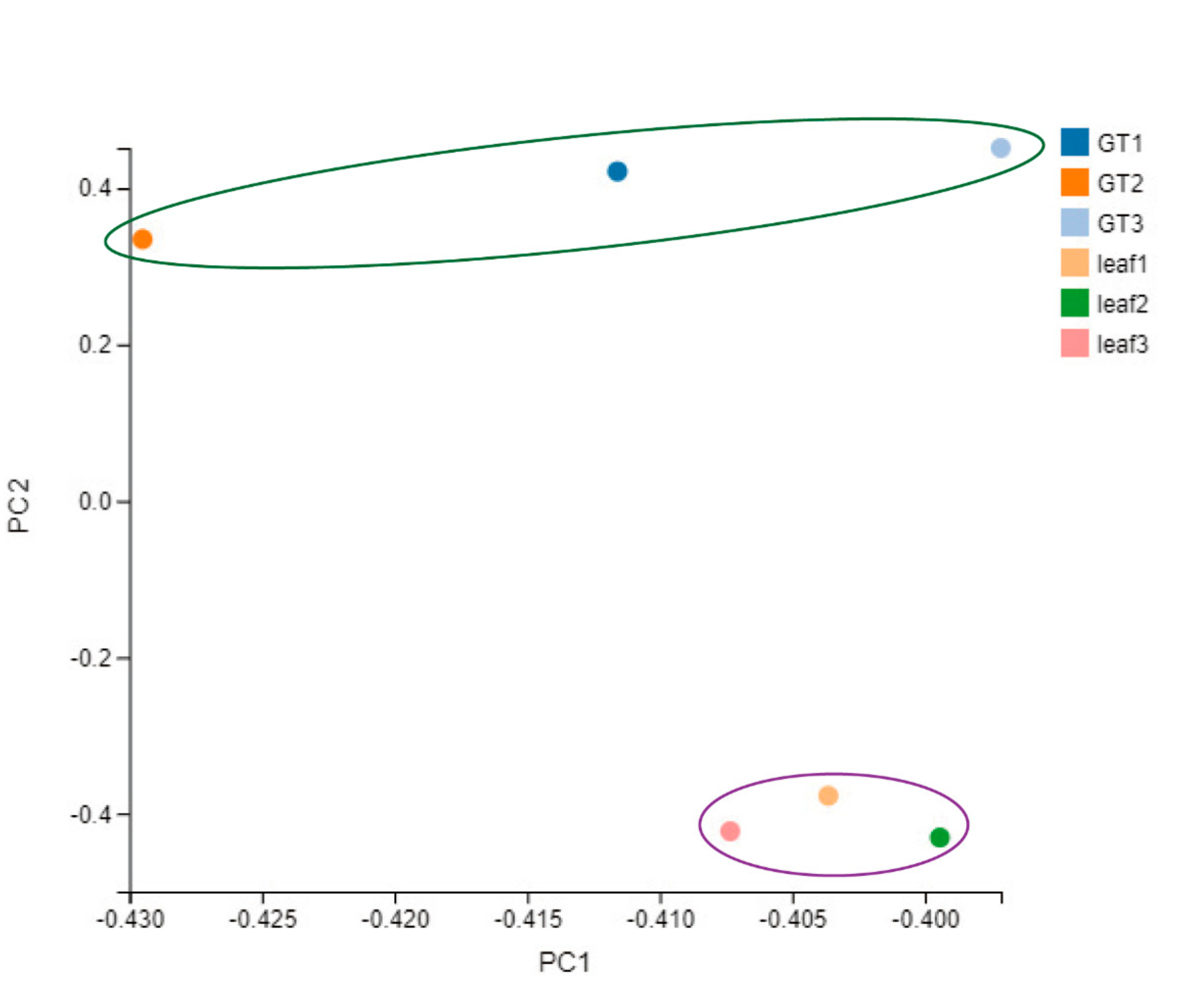
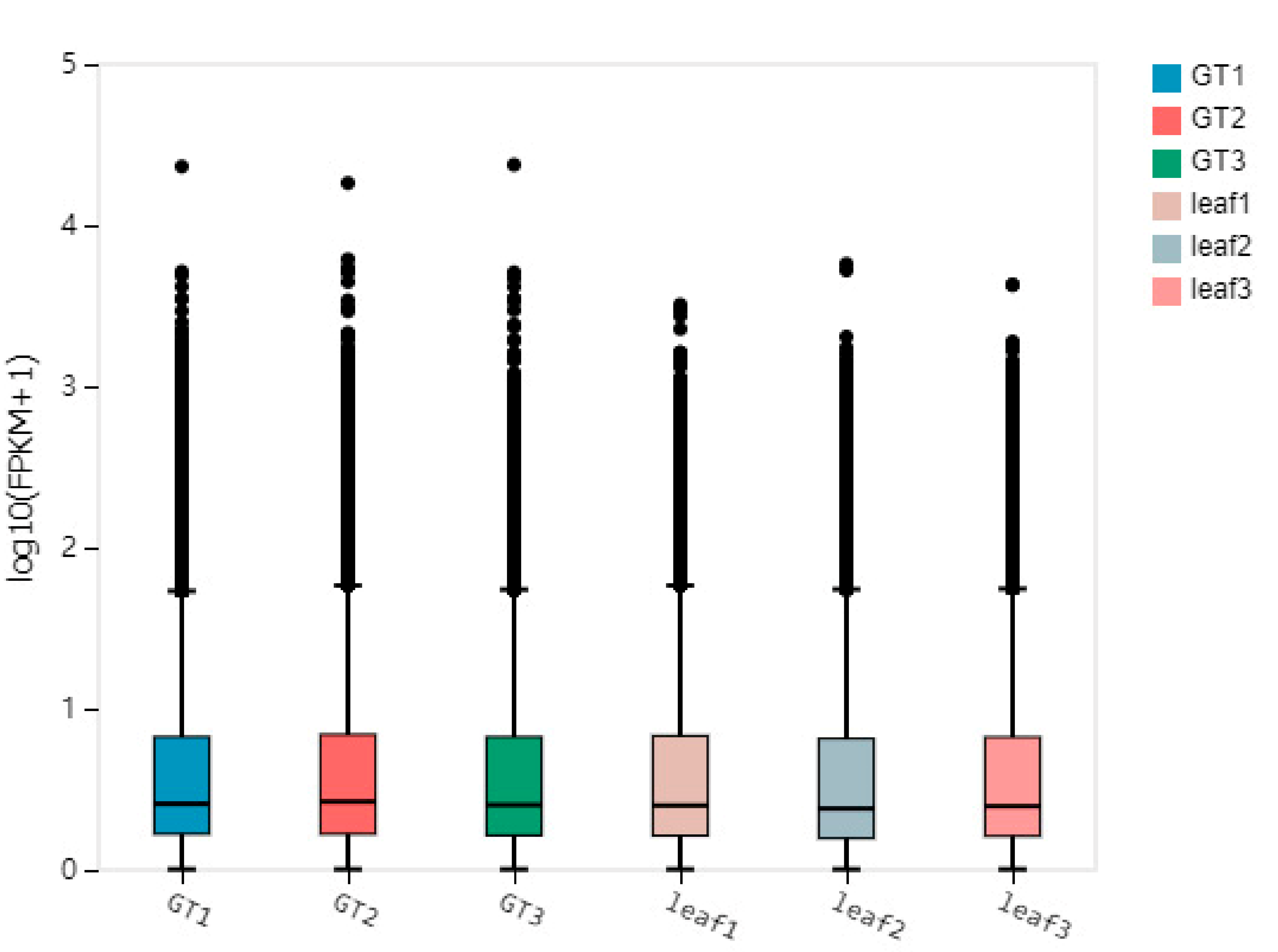
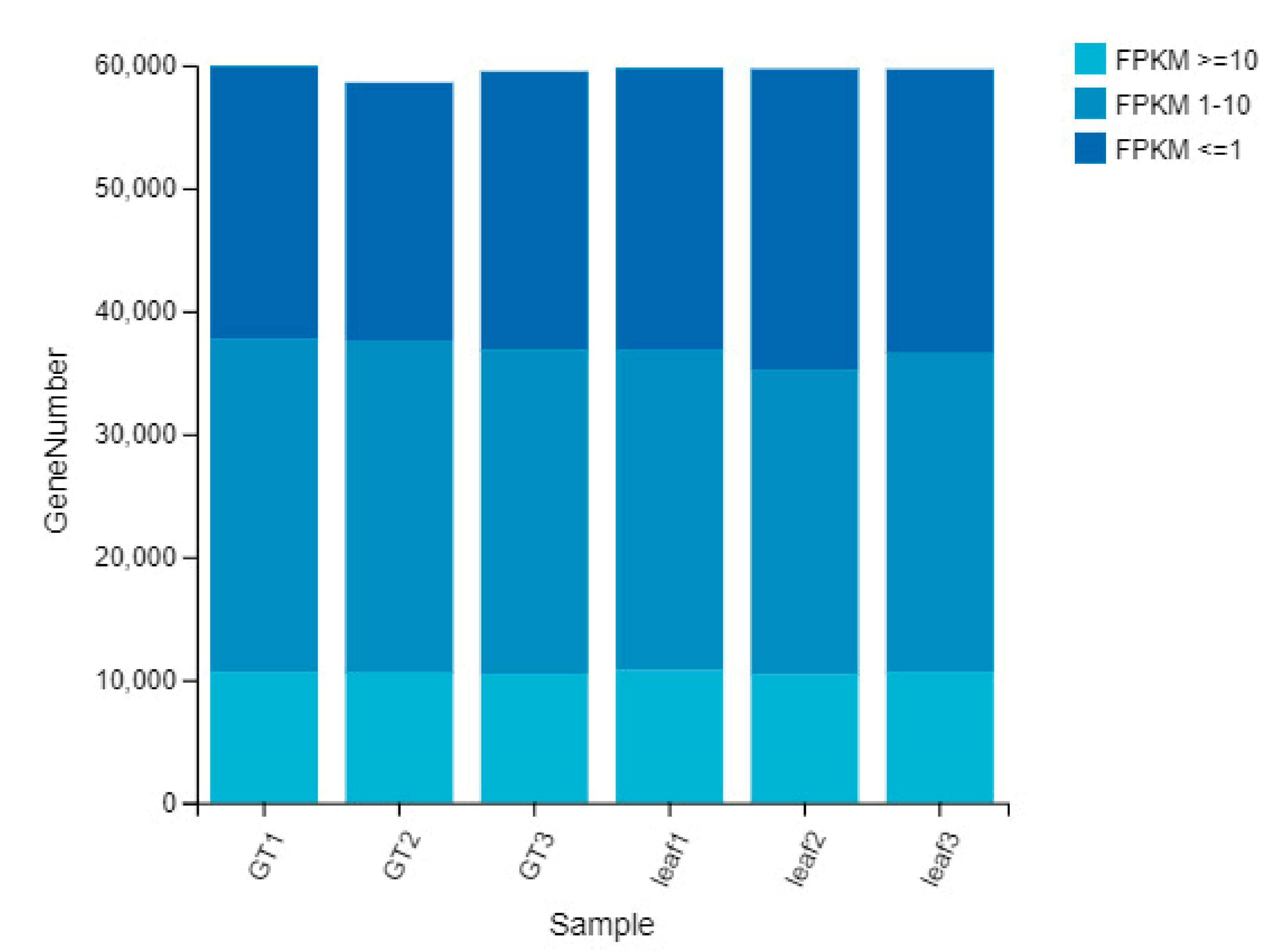
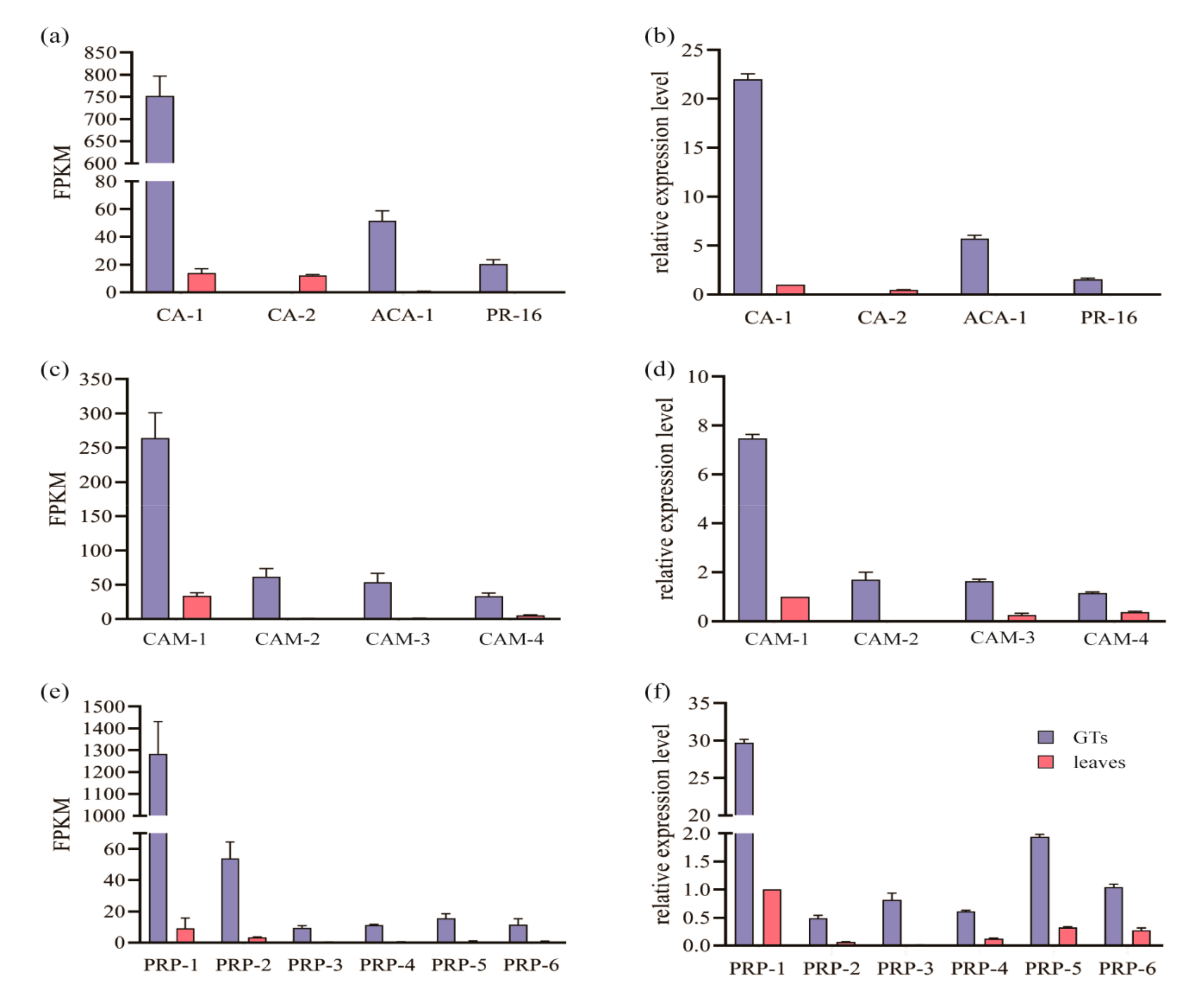
2.4. Gene Expression Pattern of the GTs
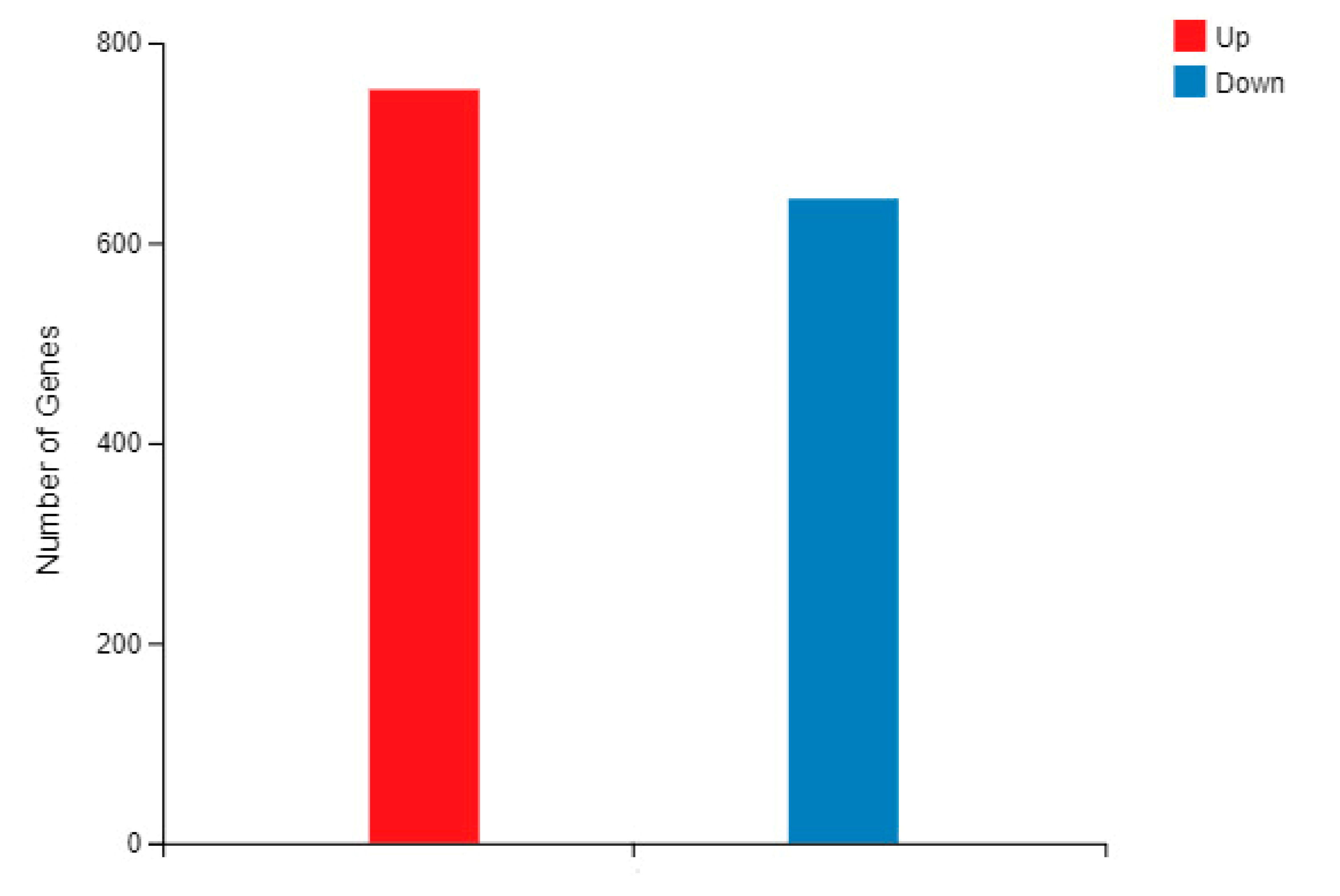
2.5. The Selectively Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Paraffin Sections
4.3. Frozen Sections
4.4. Scanning Electron Microscope (SEM)
4.5. Three-Dimensional Microscope
4.6. Transcriptome Sequencing
4.7. Gene Annotation
4.8. DEGs
4.9. Validation of Transcriptomic Data via RT-qPCR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


| Sample | Concentration (ng/μL) | Volume (μL) | Total RNA (μg) | OD260/280 | OD260/230 | RIN | 28S/18S |
|---|---|---|---|---|---|---|---|
| GT-1 | 151 | 30 | 4.53 | 1.98 | 1.18 | 8.9 | 1.5 |
| GT-2 | 79 | 15 | 1.19 | 2 | 0.61 | 9.0 | 2.9 |
| GT-3 | 143 | 15 | 2.15 | 2.04 | 0.71 | 9.3 | 2.6 |
| Tomentum-1 | 18 | 15 | 0.27 | 1.86 | 0.14 | 8.5 | 2.8 |
| Tomentum-2 | 26 | 15 | 0.39 | 1.91 | 0.69 | 8.8 | 2.8 |
| Tomentum-3 | 0.225 | 15 | 0.0034 | 2.11 | 0.08 | 2.4 | 0.0 |
| Leaf-1 | 2556 | 30 | 76.68 | 2.03 | 2.08 | 9.7 | 1.9 |
| Leaf-2 | 2168 | 30 | 65.04 | 1.98 | 1.93 | 9.0 | 1.6 |
| Leaf-3 | 3070 | 30 | 92.1 | 1.91 | 2 | 9.6 | 1.8 |
| Sample Name | Total_Raw _Read (M) | Total_Clean _Read (M) | Total_Clean _Base (Gb) | Clean_Read _q20 (%) | Clean_Read _q30 (%) | Clean_Read _Ratio (%) |
|---|---|---|---|---|---|---|
| GT-1 | 43.82 | 43.39 | 6.51 | 97.56 | 93.32 | 99.01 |
| GT-2 | 43.82 | 43.43 | 6.51 | 97.57 | 93.3 | 99.11 |
| GT-3 | 43.82 | 43.41 | 6.51 | 97.62 | 93.44 | 99.06 |
| Leaf-1 | 43.82 | 43.42 | 6.51 | 97.51 | 93.18 | 99.08 |
| Leaf-2 | 43.82 | 43.27 | 6.49 | 97.67 | 93.59 | 98.73 |
| Leaf-3 | 43.82 | 43.35 | 6.5 | 97.55 | 93.28 | 98.93 |
| Total Number | Total Length (bp) | Max Length (bp) | Min Length (bp) | Mean Length (bp) | N50 | GC% | |
|---|---|---|---|---|---|---|---|
| Unigene | 52,601 | 62,505,864 | 15,303 | 297 | 1405 | 1497 | 42.71 |
| Samples | Total Number | Total Length (bp) | Mean Length (bp) | N50 (bp) | N70 (bp) | N90 (bp) | GC (%) |
|---|---|---|---|---|---|---|---|
| GT-1 | 51,661 | 71,811,850 | 1390 | 2033 | 1466 | 732 | 40.02 |
| GT-2 | 50,439 | 65,672,100 | 1302 | 1909 | 1383 | 674 | 40.14 |
| GT-3 | 48,387 | 65,638,323 | 1356 | 1998 | 1432 | 706 | 40.19 |
| Leaf -1 | 47,472 | 67,237,102 | 1416 | 2055 | 1490 | 749 | 40.17 |
| Leaf -2 | 49,157 | 65,750,946 | 1337 | 2016 | 1439 | 684 | 40.19 |
| Leaf -3 | 49,113 | 68,843,747 | 1401 | 2060 | 1479 | 733 | 40.07 |
| Database | Numbers of Unigene | Percent (%) |
|---|---|---|
| Total | 79,032 | 100 |
| Nr | 61,027 | 77.22% |
| Nt | 52.369 | 66.26% |
| Swiss-Prot | 46,693 | 59.08% |
| KEGG | 49,677 | 62.86% |
| KOG | 48,497 | 61.36% |
| Pfam | 48,645 | 61.55 |
| GO | 46,304 | 58.59% |
| Intersection | 26,043 | 32.95% |
| Overall | 62,877 | 79.56 |
| Genes | Sense Primer | Anti-Sense Primer |
|---|---|---|
| tubulin | TGCTCTTCTGAAACACCCTTGA | CCAACTGGGCTGAGAACTAACA |
| CA-1 | CGGTGATGACAGCGTGACA | CCAGCAGGAAGAGCCATTAAC |
| CA-2 | AAGCCAAGTGCCTCTCATCA | TTACCTTCCTTGTATTCCTTCATCC |
| ACA-1 | TGCTTCTGGTTGTTGCTTCC | GTTGACTGGAGATTGGTGCTT |
| PR-16 | GATAAGAATGCCGACGACAGATT | CACAACACGCTCAAGATTGGA |
| ACM-1 | TGGTAGAGAATACTGGCGAGAT | CTCAACGGACTCAGCAACTAG |
| ACM-2 | ACGCCGAATGTGTCAGGTT | GGATAAGCATCAGATGGTTCAGAA |
| ACM-3 | TGCCTGTATCAACTATGGAGTCT | CGCTCTTCTTCTTCCTCTTCTTC |
| ACM-4 | TTCGCTTCAGCAGGAATAGGA | CCACCAACAGTTATGAGTACAAGA |
| PRP-1 | CGCAAGTCGGTTCGCATT | GCAATCATCTCTTATTCGGTGAC |
| PRP-2 | CTCCCAAAGTCAACGGCATT | CGGTTCAACGGTGAGATTCC |
| PRP-3 | TTCGCTGACATCCACTGAAGA | AGATGCTGCCACTTGAGGTT |
| PRP-4 | GAAGGAATGTGAAGCCGAAGA | TTGAAGGAGGTTGACGAATGAA |
| PRP-5 | GCCAAGGAGCAACTATGAATGA | TTCGTAGGTCGTAATCAGAATCG |
| PRP-6 | TTCAACGACCAGCACGAGTA | CCTTCATTCCTCTTCAGCATCTC |
References
- McAdam, E.L.; Brodribb, T.J.; McAdam, S.A.M. Does ozone increase ABA levels by non-enzymatic synthesis causing stomata to close? Plant Cell Environ. 2017, 40, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Bosu, P.P.; Wagner, M.R. Effects of induced water stress on leaf trichome density and foliar nutrients of three elm (Ulmus) species: Implications for resistance to the elm leaf beetle. Environ. Èntomol. 2007, 36, 595–601. [Google Scholar] [CrossRef] [Green Version]
- Delaveau, P. Biology and chemistry of plant trichomes. Comp. Biochem. Physiol. Part A Physiol. 1985, 81, 455. [Google Scholar] [CrossRef]
- Fahn, A. Structure and function of secretory cells. Adv. Bot. Res. 2000, 31, 37–75. [Google Scholar]
- Ascensão, L.; Mota, L.; Castro, M.D.M. Glandular Trichomes on the Leaves and Flowers of Plectranthus ornatus: Morphology, Distribution and Histochemistry. Ann. Bot. 1999, 84, 437–447. [Google Scholar] [CrossRef]
- Payne, W.W. A Glossary of Plant Hair Terminology. Brittonia 1978, 30, 239–255. [Google Scholar] [CrossRef]
- Glas, J.J.; Schimmel, B.C.J.; Alba, J.M.; Escobar-Bravo, R.; Schuurink, R.C.; Kant, M.R. Plant Glandular Trichomes as Targets for Breeding or Engineering of Resistance to Herbivores. Int. J. Mol. Sci. 2012, 13, 17077–17103. [Google Scholar] [CrossRef] [Green Version]
- Werker, E. Trichome diversity and development. Adv. Bot. Res. 2000, 31, 1–35. [Google Scholar] [CrossRef]
- Sharma, S.; Sangwan, N.S.; Sangwan, R.S. Developmental process of essential oil glandular trichome collapsing in menthol mint (Mentha arvensis). Curr. Sci. 2003, 84, 544–550. [Google Scholar] [CrossRef]
- Xu, J.; Van Herwijnen, Z.O.; Dräger, D.; Sui, C.; Haring, M.A.; Schuurink, R. SlMYC1 Regulates Type VI Glandular Trichome Formation and Terpene Biosynthesis in Tomato Glandular Cells. Plant Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef] [Green Version]
- Ambrósio, S.R.; Oki, Y.; Heleno, V.C.G.; Chaves, J.; Nascimento, P.G.B.D.; Lichston, J.E.; Constantino, M.G.; Varanda, E.M.; Da Costa, F.B. Constituents of glandular trichomes of Tithonia diversifolia: Relationships to herbivory and antifeedant activity. Phytochemistry 2008, 69, 2052–2060. [Google Scholar] [CrossRef] [PubMed]
- Weathers, P.J.; Arsenault, P.R.; Covello, P.S.; McMickle, A.; Teoh, K.H.; Reed, D. Artemisinin production in Artemisia annua: Studies in planta and results of a novel delivery method for treating malaria and other neglected diseases. Phytochem. Rev. 2010, 10, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Kelsey, R.G. Glandular Trichomes: A Helpful Taxonomic Character for Artemisia nova (Black Sagebrush). J. Range Manag. 1984, 37, 370–372. [Google Scholar] [CrossRef]
- Rong, X.; Lei, Z.; Xue, D.; Gue, J.; Deng, J. Study on relationship between the compoundleaf type of Rhus potaninii Maximwith the yield of gall. Hubei For. Sci. Technol. 1995, 3, 11. [Google Scholar]
- Chen, X.-M.; Feng, Y. An Introduction to Resource Entomology; The Science Press: Beijing, China, 2009. [Google Scholar]
- Ascensão, L.; Marques, N.; Pais, M.S. Glandular Trichomes on Vegetative and Reproductive Organs of Leonotis leonurus (Lamiaceæ). Ann. Bot. 1995, 75, 619–626. [Google Scholar] [CrossRef]
- Uchiyama, T.; Ogasawara, N. Disappearance of the Cuticle and Wax in Outermost Layer of Callus Cultures and Decrease of Protective Ability against Microorganisms. Agric. Biol. Chem. 1977, 41, 1401–1405. [Google Scholar] [CrossRef]
- Clifford, S.; Arndt, S.; Popp, M.; Jones, H. Mucilages and polysaccharides in Ziziphus species (Rhamnaceae): Localization, composition and physiological roles during drought-stress. J. Exp. Bot. 2002, 53, 131–138. [Google Scholar] [CrossRef]
- Fahn, A. Secretory tissues in vascular plants. New Phytol. 1988, 108, 229–257. [Google Scholar] [CrossRef]
- Modenesi, P.; Serrato-Valenti, G.; Bruni, A. Development and Secretion of Clubbed Trichomes in Thymus vulgaris L. Flora Morphol. Distrib. Funct. Ecol. Plants 1984, 175, 211–219. [Google Scholar] [CrossRef]
- Lattanzio, V.; Lattanzio, V.M.T.; Cardinali, A.; Imperato, F. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. Phytochemistry 2006, 37, 23–67. [Google Scholar]
- Rong, Z. Cyanide. Available online: https://baike.baidu.com/item/%E6%B0%B0%E5%8C%96%E7%89%A9/1136704?fr=aladdin (accessed on 10 November 2020).
- Xue, Z.; Ye, R.; Yun, T.; Wei, Z.; Wang, J.; Han, Y.; Huang, Y.; Yuan, L. The Plant Morphology and Total Polysaccharides Content in the Leaves of Panax quinquefolius L. Grown in Canada. Med. Plant 2015, 6, 25–27. [Google Scholar]
- Lacchia, A.P.S.; Tölke, E.E.D.; Carmello-Guerreiro, S.M.; Ascensão, L.; Demarco, D. Foliar colleters in Anacardiaceae: First report for the family. Botany 2016, 94, 337–346. [Google Scholar] [CrossRef]
- Jin, J.; Panicker, D.; Wang, Q.; Kim, M.J.; Liu, J.; Yin, J.-L.; Wong, L.; Jang, I.-C.; Chua, N.-H.; Sarojam, R. Next generation sequencing unravels the biosynthetic ability of Spearmint (Mentha spicata) peltate glandular trichomes through comparative transcriptomics. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, S.; Sato, F. Plant Pathogenesis-Related Proteins: Molecular Mechanisms of Gene Expression and Protein Function. J. Biochem. 1999, 125, 1–8. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Galdon-Armero, J.; Fullana-Pericàs, M.; Mulet, P.A.; Conesa, M.A.; Martin, C.; Galmes, J. The ratio of trichomes to stomata is associated with water use efficiency inSolanum lycopersicum(tomato). Plant J. 2018, 96, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Wagner, G.J. Secreting Glandular Trichomes: More than Just Hairs. Plant Physiol. 1991, 96, 675–679. [Google Scholar] [CrossRef] [Green Version]
- Jensen, W.A. Botanical Histochemistry; W.H. Freeman: San Francisco, CA, USA, 1962; pp. 74–99. [Google Scholar]
- Pizzolato, P.; Lillie, R.D. Mayer’s Tannic Acid-Ferric Chloride Stain for Mucins. J. Histochem. Cytochem. 1973, 21, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Cain, A.J. The Use of Nile Blue in the Examination of Lipoids. J. Cell Sci. 1947, s3-88, 383–392. [Google Scholar] [CrossRef]
- McCracken, E.; Johansen, D.A. Plant Microtechnique. Trans. Am. Microsc. Soc. 1940, 59, 405. [Google Scholar] [CrossRef]
- Pearse, A.G.E. Histochemistry, Theoretical and Applied. J. Anat. 1980, 87, 46–98. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Amit, I. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schena, L.; Nigro, F.; Ippolito, A.; Gallitelli, D. Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. Eur. J. Plant Pathol. 2004, 110, 893–908. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Q.; Bashir, N.H.; Wu, H.-X.; Wang, W.; Zhang, J.; Cui, Y.; Chen, H. Structure, Distribution, Chemical Composition, and Gene Expression Pattern of Glandular Trichomes on the Leaves of Rhus potaninii Maxim. Int. J. Mol. Sci. 2021, 22, 7312. https://doi.org/10.3390/ijms22147312
Lu Q, Bashir NH, Wu H-X, Wang W, Zhang J, Cui Y, Chen H. Structure, Distribution, Chemical Composition, and Gene Expression Pattern of Glandular Trichomes on the Leaves of Rhus potaninii Maxim. International Journal of Molecular Sciences. 2021; 22(14):7312. https://doi.org/10.3390/ijms22147312
Chicago/Turabian StyleLu, Qin, Nawaz Haider Bashir, Hai-Xia Wu, Weiwei Wang, Jinwen Zhang, Yongzhong Cui, and Hang Chen. 2021. "Structure, Distribution, Chemical Composition, and Gene Expression Pattern of Glandular Trichomes on the Leaves of Rhus potaninii Maxim" International Journal of Molecular Sciences 22, no. 14: 7312. https://doi.org/10.3390/ijms22147312






