Selective Antifungal Activity and Fungal Biofilm Inhibition of Tryptophan Center Symmetrical Short Peptide
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Activity
2.2. Structure Variability of the Peptides
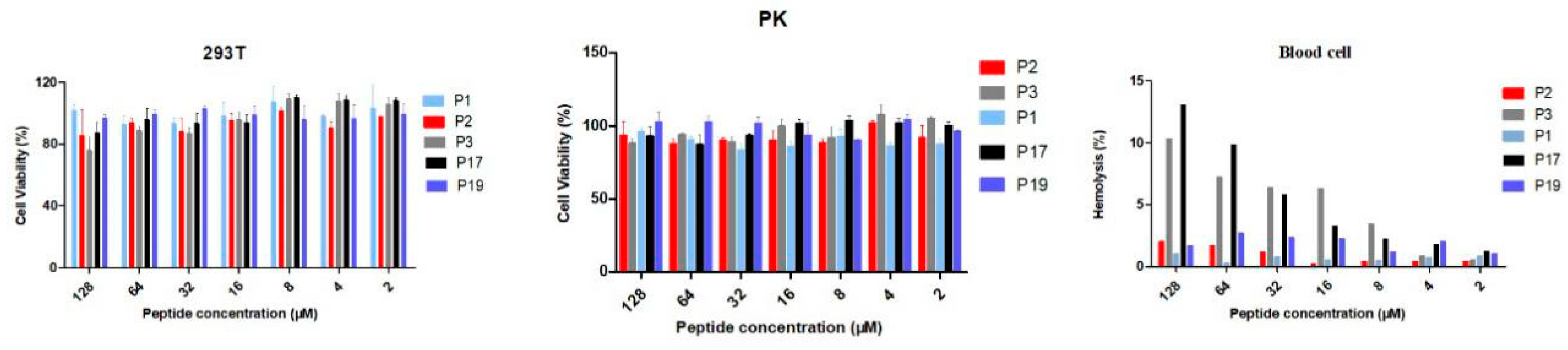
2.3. Biocompatibility of the Peptides
2.4. Sensitivity upon Salt and Acid Condition
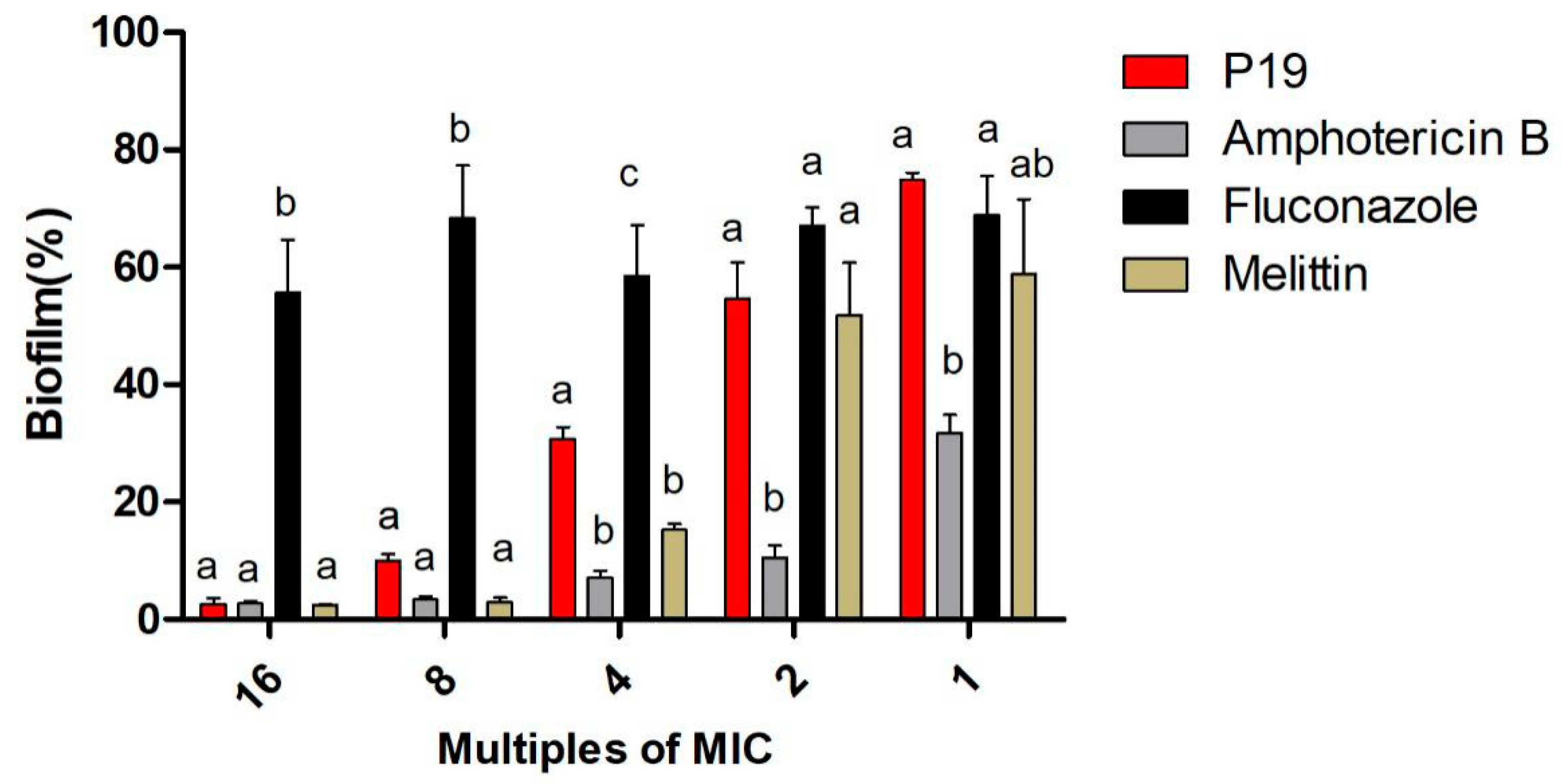
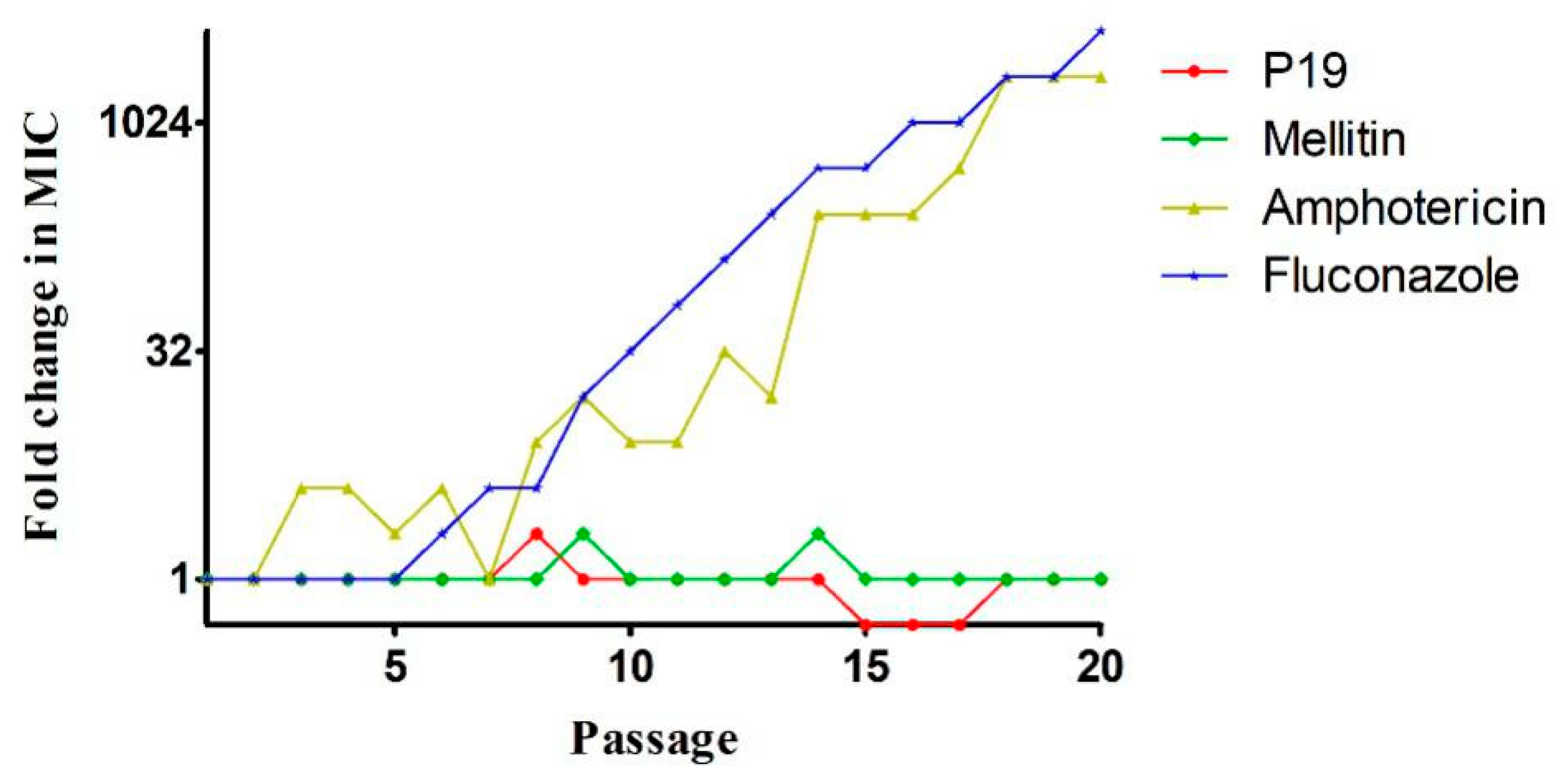
2.5. Drug-Resistance
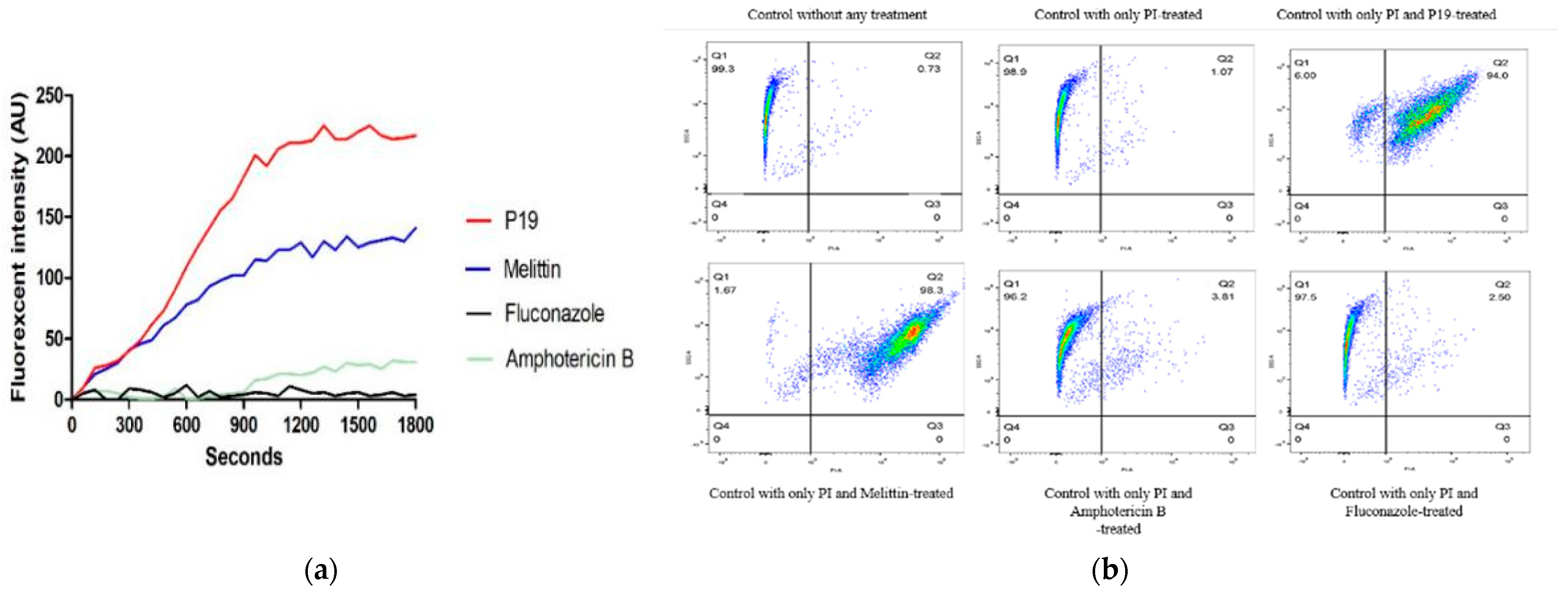
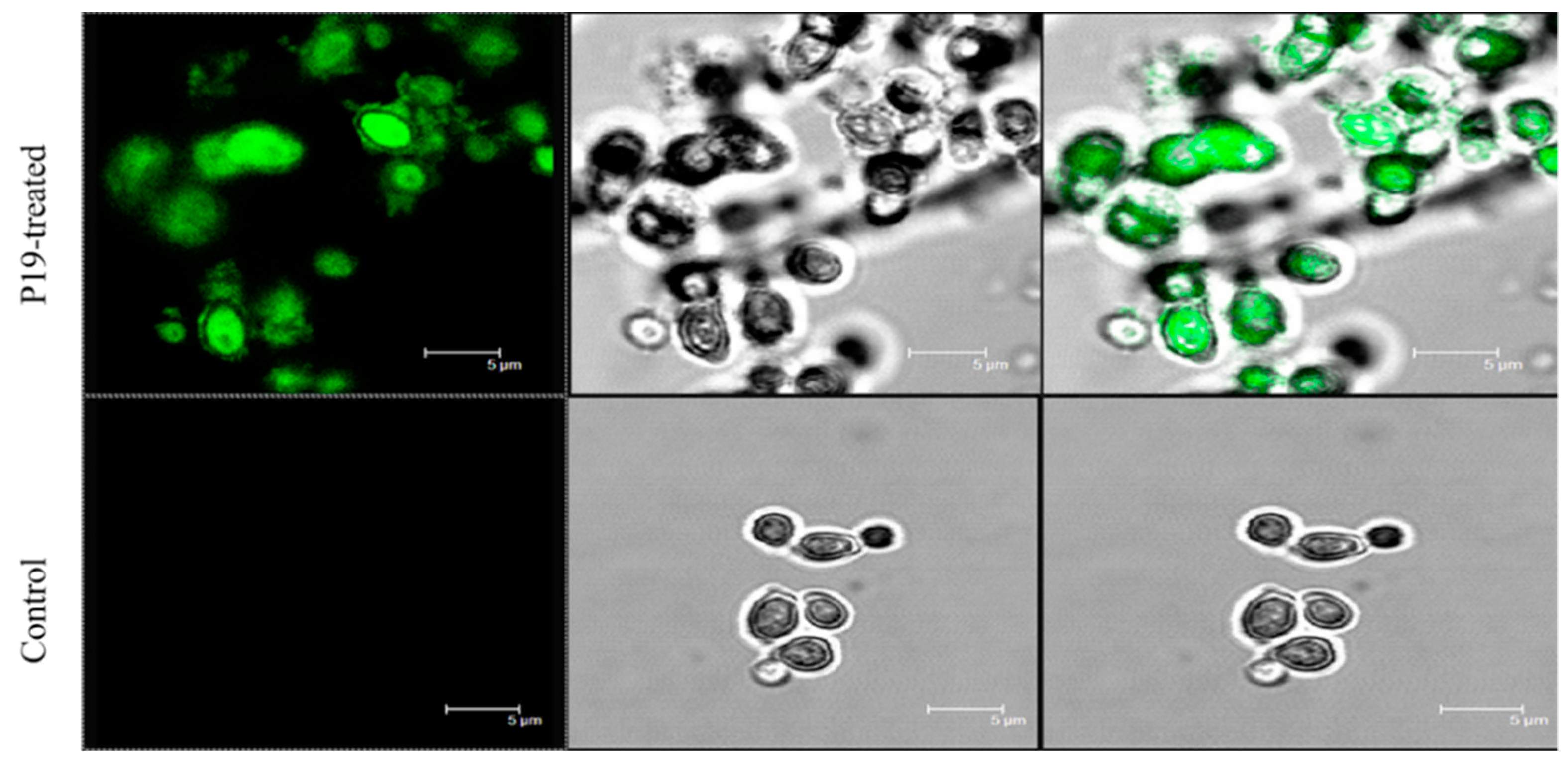
2.6. Membrane Permeabilization and Integrity
2.7. Membrane Binding Affinity
3. Discussion
4. Materials and Methods
4.1. Bacteria and Fungi
4.2. Peptide Synthesis
4.3. Circular Dichroism (CD) Measurements
4.4. Antimicrobial Assays
4.5. Cytotoxicity Assays
4.6. Salt and Acid Sensitivity Assays
4.7. Drug-Resistance
4.8. Membrane Permeabilization and Integrity Assays
4.8.1. Cytoplasmic Membrane Depolarization Assay
4.8.2. Flow Cytometer Assay
4.8.3. SEM, TEM and CLSM Characterization
4.9. Membrane Binding Affinity Assays
4.9.1. Whole Cell ELISA
4.9.2. Protein–Lipid Overlay Assay
4.10. Binding Affinity to LPS or LTA
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMPs | Antimicrobial peptides |
| LPS | Lipopolysaccharides |
| LTA | Lipoteichoic acids |
| MALDI-TOF-MS | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| RP-HPLC | Reverse-phase high performance liquid chromatography |
| SEM | Scanning electron microscopy |
| TEM | Transmission electron microscopy |
| CLSM | Confocal laser scanning microscopy |
| CD | Circular dichroism |
| PI | Phosphatidylinositol |
| PIP | Phosphatidylinositol (4,5)bisphosphate |
| PS | Phosphatidylserine |
| PG | Phosphatidylglycerol |
| FITC | Fluorescein Isothiocyanate |
| PBS | Phosphate-buffered saline |
| PBST | Phosphate-buffered saline with 0.05% tween 20 |
| TFE | Trifluoroethanol |
| SDS | Sodium lauryl sulfate |
| MICs | The minimum inhibitory concentrations |
| MTT | 3-[4,5-dimethylthiozol-2-yl]-2,5-diphenyltetrazolium bromide |
| YPDA | Yeast Peptone Dextrose Agar |
| MOPs | Morpholinepropanesulfonic acid |
| MHB | Mueller-Hinton Broth |
| DiSC3–5 | 3,3-dipropylthiadicarbocyanine |
| CDI | Clostridium difficile infection |
| FMT | Fecal microbiota transplantation |
| APD | The Antimicrobial Peptide Database |
| TMP | Tetramethylbenzidine membrane peroxidase |
References
- Muñoz, A.; Gandía, M.; Harries, E.; Carmona, L.; Read, N.D.; Marcos, J.F. Understanding the Mechanism of Action of Cell-Penetrating Antifungal Peptides Using the Rationally Designed Hexapeptide PAF26 as a Model. Fungal Biol. Rev. 2013, 26, 146–155. [Google Scholar] [CrossRef]
- Zuo, T.; Wong, S.H.; Cheung, C.P.; Lam, K.; Lui, R.; Cheung, K.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Wu, J.C.Y.; et al. Gut Fungal Dysbiosis Correlates with Reduced Efficacy of Fecal Microbiota Transplantation in Clostridium Difficile Infection. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Ren, B.; He, J.; Peng, X.; Guo, Q.; Zheng, L.; Li, J.; Dai, H.; Chen, V.; Zhang, L.; et al. Candida Albicans Promotes Tooth Decay by Inducing Oral Microbial Dysbiosis. ISME J. 2020, 15, 894–908. [Google Scholar] [CrossRef] [PubMed]
- Faruck, M.O.; Yusof, F.; Chowdhury, S. An Overview of Antifungal Peptides Derived from Insect. Peptides 2016, 80, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide Emergence of Resistance to Antifungal Drugs Challenges Human Health and Food Security. Science 2018, 742, 739–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLellan, C.A.; Vincent, B.M.; Solis, N.V.; Lancaster, A.K.; Sullivan, L.B.; Hartland, C.L.; Youngsaye, W.; Filler, S.G.; Whitesell, L. Inhibiting Mitochondrial Phosphate Transport as an Unexploited Antifungal Strategy. Nat. Chem. Biol. 2018, 14, 135–141. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Song, D.W.; Kim, S.H.; Kim, H.H.; Lee, K.H.; Ki, C.S.; Park, Y.H. Multi Biofunction of Antimicrobial Peptide-Immobilized Silk Fibroin Nanofiber Membrane: Implications for Wound Healing. Acta Biomater. 2016, 39, 146–155. [Google Scholar] [CrossRef]
- Muñoz, A.; Read, N.D. Live-Cell Imaging and Analysis Shed Light on the Complexity and Dynamics of Antimicrobial Peptide Action. Front. Immunol. 2012, 3, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Khara, J.S.; Obuobi, S.; Wang, Y.; Hamilton, M.S.; Robertson, B.D.; Newton, S.M.; Yang, Y.Y.; Langford, P.R.; Ee, P.L.R. Disruption of Drug-Resistant Biofilms Using de Novo Designed Short α-Helical Antimicrobial Peptides with Idealized Facial Amphiphilicity. Acta Biomater. 2017, 57, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Maraming, P. The Cationic Cell - Penetrating KT2 Peptide Promotes Cell Membrane Defects and Apoptosis with Autophagy Inhibition in Human HCT 116 Colon Cancer Cells. Cell. Physiol. 2019, 234, 1–14. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Shan, A.; Feng, X. Avian host defense cathelicidins: Structure, expression, biological functions, and potential therapeutic applications. Poult. Sci. 2020, 99, 6434–6445. [Google Scholar] [CrossRef]
- Wang, J.; Chou, S.; Yang, Z.; Yang, Y.; Wang, Z.; Song, J.; Dou, X.; Shan, A. Combating Drug-Resistant Fungi with Novel Imperfectly Amphipathic Palindromic Peptides. J. Med. Chem. 2018, 61, 3889–3907. [Google Scholar] [CrossRef]
- Chou, S.; Shao, C.; Wang, J.; Shan, A.; Xu, L.; Dong, N.; Li, Z. Short, Multiple-Stranded β-Hairpin Peptides Have Antimicrobial Potency with High Selectivity and Salt Resistance. Acta Biomater. 2016, 30, 78–93. [Google Scholar] [CrossRef]
- Shukla, A.; Fleming, K.E.; Chuang, H.F.; Chau, T.M.; Loose, C.R.; Stephanopoulos, G.N.; Hammond, P.T. Controlling the Release of Peptide Antimicrobial Agents from Surfaces. Biomaterials 2010, 31, 2348–2357. [Google Scholar] [CrossRef]
- De Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; De Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; Van Der Heijde, T.; Boekema, B.K.; et al. The Antimicrobial Peptide SAAP-148 Combats Drug-Resistant Bacteria and Biofilms. Sci. Transl. Med. 2018, 10, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camele, I.; Elshafie, H.S.; Caputo, L.; Sakr, S.H.; De Feo, V. Bacillus mojavensis: Biofilm formation and biochemical investigation of its bioactive metabolites. J. Biol. Res. 2019, 92, 39–45. [Google Scholar] [CrossRef]
- Ng, S.M.S.; Yap, J.M.; Lau, Q.Y.; Ng, F.M.; Ong, E.H.Q.; Barkham, T.; Teo, J.W.P.; Alfatah, M.; Kong, K.W.; Hoon, S.; et al. Structure-Activity Relationship Studies of Ultra-Short Peptides with Potent Activities against Fluconazole-Resistant Candida Albicans. Eur. J. Med. Chem. 2018, 150, 479–490. [Google Scholar] [CrossRef]
- Jagadish, K.; Gould, A.; Borra, R.; Majumder, S.; Mushtaq, Z.; Shekhtman, A.; Camarero, J.A. Recombinant Expression and Phenotypic Screening of a Bioactive Cyclotide Against α-Synuclein-Induced Cytotoxicity in Baker’s Yeast. Angew. Chemie Int. Ed. 2015, 54, 8390–8394. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, K.A.; Kelly, M.A.; Li, K.; Cambray, S.; Hosseini, A.S.; Van Opijnen, T.; Gao, J. Phage Display of Dynamic Covalent Binding Motifs Enables Facile Development of Targeted Antibiotics. J. Am. Chem. Soc. 2018, 140, 6137–6145. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Wang, J.; Shang, L.; Akhtar, M.U.; Wang, Z.; Shi, B.; Feng, X.; Shan, A. Short, Symmetric-Helical Peptides Have Narrow-Spectrum Activity with Low Resistance Potential and High Selectivity. Biomater. Sci. 2019, 7, 2394–2409. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Li, Q.; Nina, Z.; Shang, L.; Li, J.; Li, J.; Wang, Z.; Shan, A. Peptides With Triplet-Tryptophan-Pivot Promoted Pathogenic Bacteria Membrane Defects. Front. Microbiol. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiradharma, N.; Khoe, U.; Hauser, C.A.E.; Seow, S.V.; Zhang, S.; Yang, Y.Y. Synthetic Cationic Amphiphilic α-Helical Peptides as Antimicrobial Agents. Biomaterials 2011, 32, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Tan, P.; Zhu, Y.; Shao, C.; Shan, A.; Li, L. Highly Stabilized α -Helical Coiled Coils Kill Gram-Negative Bacteria by Multi-Complementary Mechanisms under Acidic Condition. Biol. Med. Appl. Mater. Interfaces 2019, 11, 22113–22128. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Wang, Z.; Chou, S.; Zhang, L.; Shan, A.; Jiang, J. Antibacterial Activities and Molecular Mechanism of Amino-Terminal Fragments from Pig Nematode Antimicrobial Peptide CP-1. Chem. Biol. Drug Des. 2018, 91, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Wang, C.; Zhang, T.; Zhang, L.; Xue, C.; Feng, X.; Bi, C.; Shan, A. Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria. Int. J. Mol. Sci. 2019, 20, 3954. [Google Scholar] [CrossRef] [Green Version]
- Stone, T.A.; Cole, G.B.; Ravamehr-lake, D.; Nguyen, Q.; Khan, F.; Sharpe, S.; Deber, C.M.; Stone, T.A.; Cole, G.B.; Ravamehr-lake, D.; et al. Positive Charge Patterning and Hydrophobicity of Membrane-Active Antimicrobial Peptides as Determinants of Activity, Toxicity, and Pharmacokinetic Stability. J. Med. Chem. 2019, 62, 6276–6286. [Google Scholar] [CrossRef]
- Gopal, R.; Seo, C.H.; Song, P.I.; Park, Y. Effect of Repetitive Lysine Tryptophan Motifs on the Bactericidal Activity of Antimicrobial Peptides. Amino Acids 2013, 44, 645–660. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.; Chou, S.; Li, J.; Xue, C.; Li, X.; Cheng, B.; Shan, A.; Xu, L. Short Symmetric-End Antimicrobial Peptides Centered on β-Turn Amino Acids Unit Improve Selectivity and Stability. Front. Microbiol. 2018, 9, 2832. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Zhu, Y.; Jian, Q.; Lai, Z.; Tan, P.; Li, G.; Shan, A. Cross-Strand Interaction, Central Bending, and Sequence Pattern Act as Biomodulators of Simplified β-Hairpin Antimicrobial Amphiphiles. Small 2021, 17, 2003899. [Google Scholar] [CrossRef]
- Li, Q.; Li, J.; Yu, W.; Wang, Z.; Li, J.; Feng, X.; Wang, J.; Shan, A. De novo design of a pH-triggered self-assembled β-hairpin nanopeptide with the dual biological functions for antibacterial and entrapment. J. Nanobiotechnol. 2021, 19, 1–19. [Google Scholar] [CrossRef]
- Wang, C.; Shao, C.; Fang, Y.; Wang, J.; Dong, N.; Shan, A.. Binding loop of sunflower trypsin inhibitor 1 serves as a design motif for proteolysis-resistant antimicrobial peptides. Acta Biomater. 2021, 124, 254–269. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-Positive Bacterial Cell Envelopes: The Impact on the Activity of Antimicrobial Peptides. Biochim. Biophys. Acta Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef] [Green Version]
- Järvå, M.; Phan, T.K.; Lay, F.T.; Caria, S.; Kvansakul, M.; Hulett, M.D. Human-defensin 2 Kills Candida Albicans through Phosphatidylinositol 4,5Bisphosphate–Mediated Membrane Permeabilization. Sci. Adv. 2018, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löffler, J.; Einsele, H.; Hebart, H.; Schumacher, U.; Hrastnik, C.; Daum, G. Phospholipid and Sterol Analysis of Plasma Membranes of Azole-Resistant Candida albicans Strains. FEMS Microbiol. Lett. 2000, 185, 59–63. [Google Scholar] [CrossRef]
- White, D.A.; Lennarz, W.J.; Schnaitman, C.A. Distribution of Lipids in the Wall and Cytoplasmic Membrane Subfractions of the Cell Envelope of Escherichia coli. J. Bacteriol. 1972, 109, 686–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, N.N.; Bayer, A.S. Correlation of Cell Membrane Lipid Profiles with Daptomycin Resistance in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 1082–1085. [Google Scholar] [CrossRef] [Green Version]
- Ran, S.; Thorpe, P.E. Phosphatidylserine Is a Marker of Tumor Vasculature and a Potential Target for Cancer Imaging and Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2002, 54, 1479–1484. [Google Scholar] [CrossRef]
- Fidler, I.J.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated Expression of Phosphatidylseine in the Outer Membrane Leaflet of Human Tumor Cells and Recognition by Activated Human Blood Monocytes. Cancer Res. 1991, 51, 3062–3066. [Google Scholar]
- Yamazaki, D.; Kurisu, S.; Takenawa, T. Regulation of Cancer Cell Motility through Actin Reorganzation. Cancer Sci. 2005, 96, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Aoki, M.; Yano, Y.; Matsuzaki, K. Interaction of Antimicrobial Peptide Magainin 2 with Gangliosides as a Target for Human Cell Binding. Biochemistry 2012, 51, 10229–10235. [Google Scholar] [CrossRef]
- Saporito, P.; Biljana, M.; Løbner, A.; Håvard, O. Antibacterial Mechanisms of GN-2 Derived Peptides and Peptoids against Escherichia coli. Biopolymers 2019, 110, e23275. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Li, W.; Tan, P.; Shan, A.; Dou, X.; Ma, D.; Liu, C. Symmetrical Modification of Minimized Dermaseptins to Extend the Spectrum of Antimicrobials with Endotoxin Neutralization Potency. Int. J. Mol. Sci. 2019, 20, 1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]

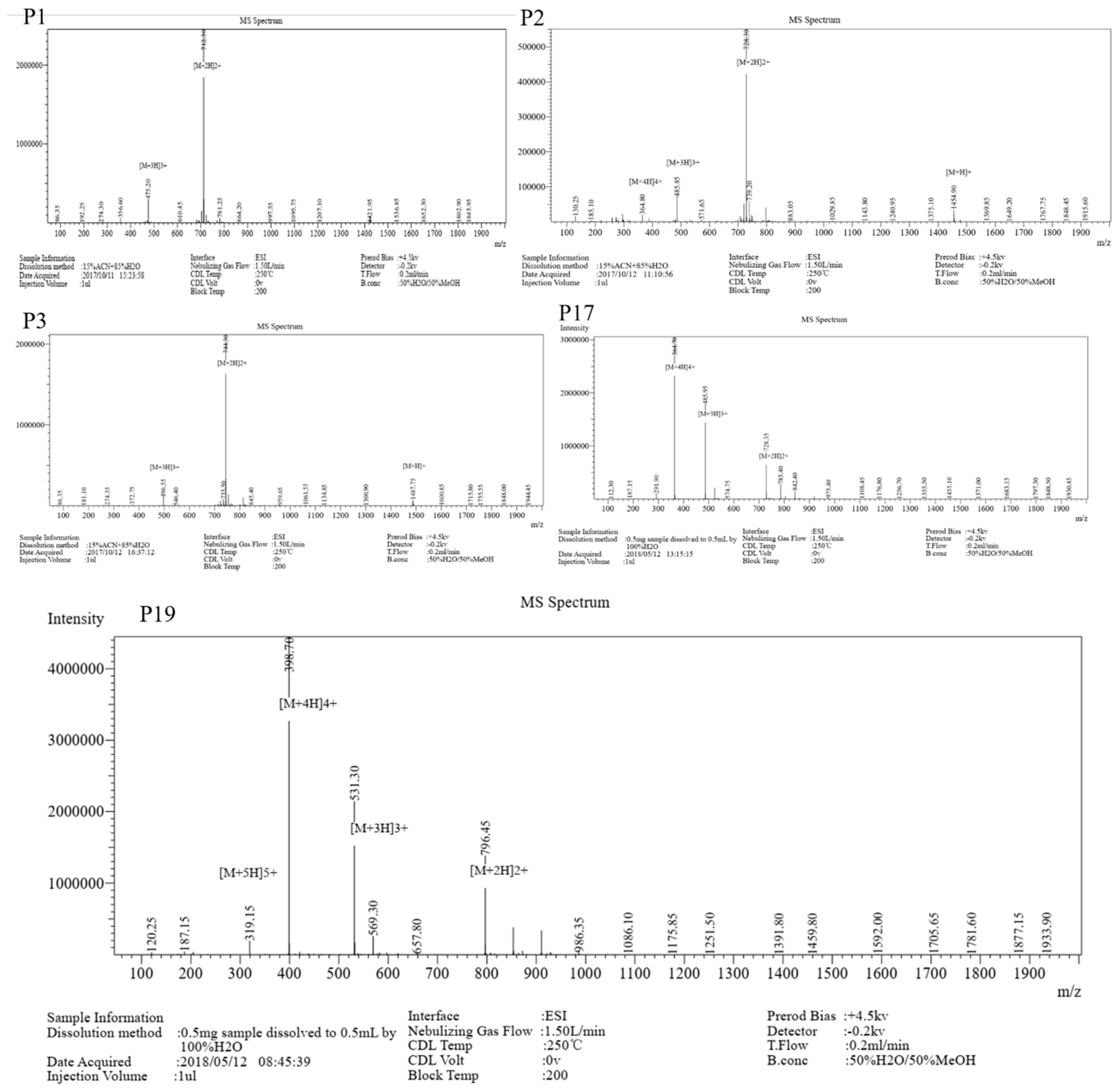
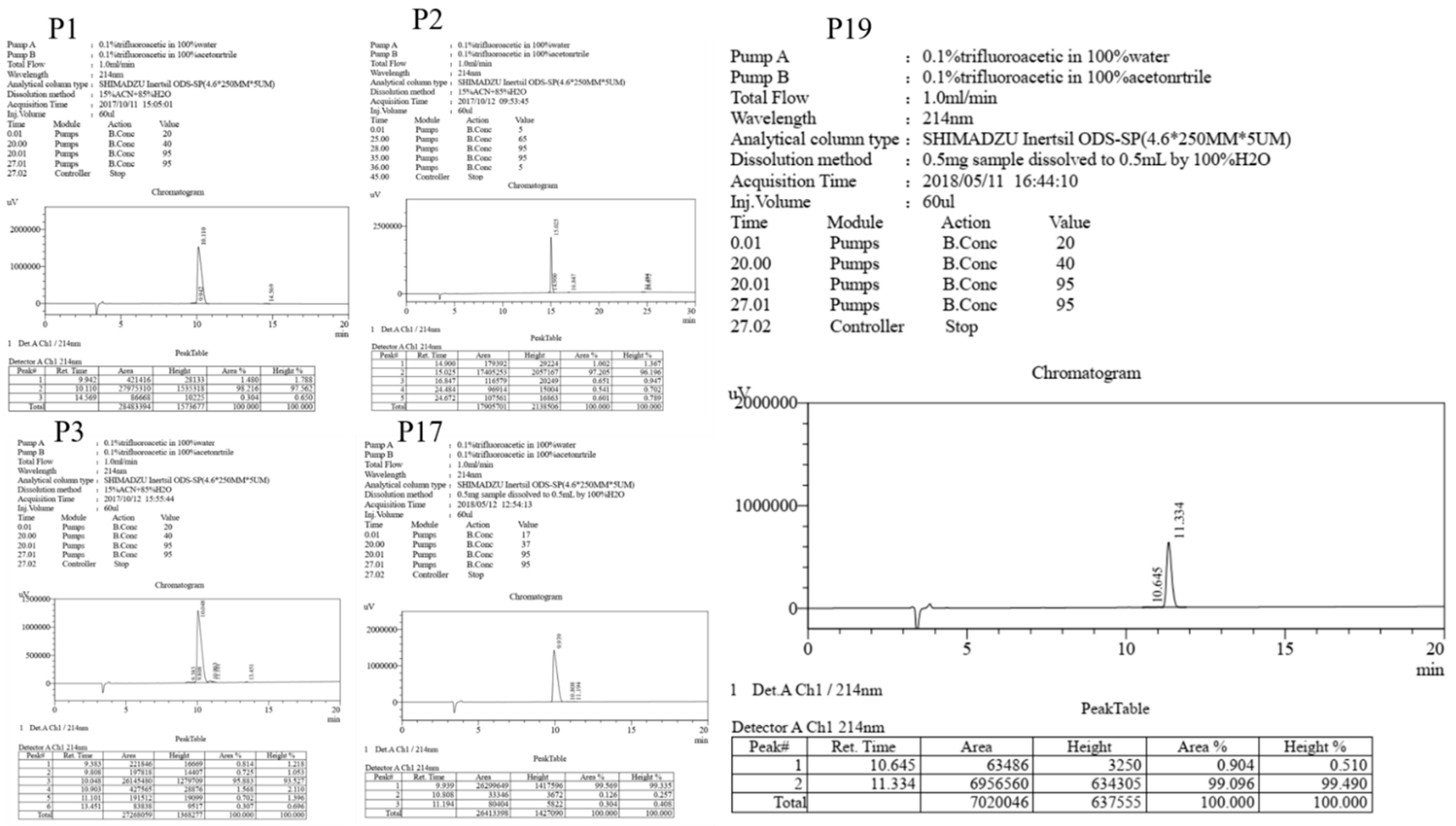
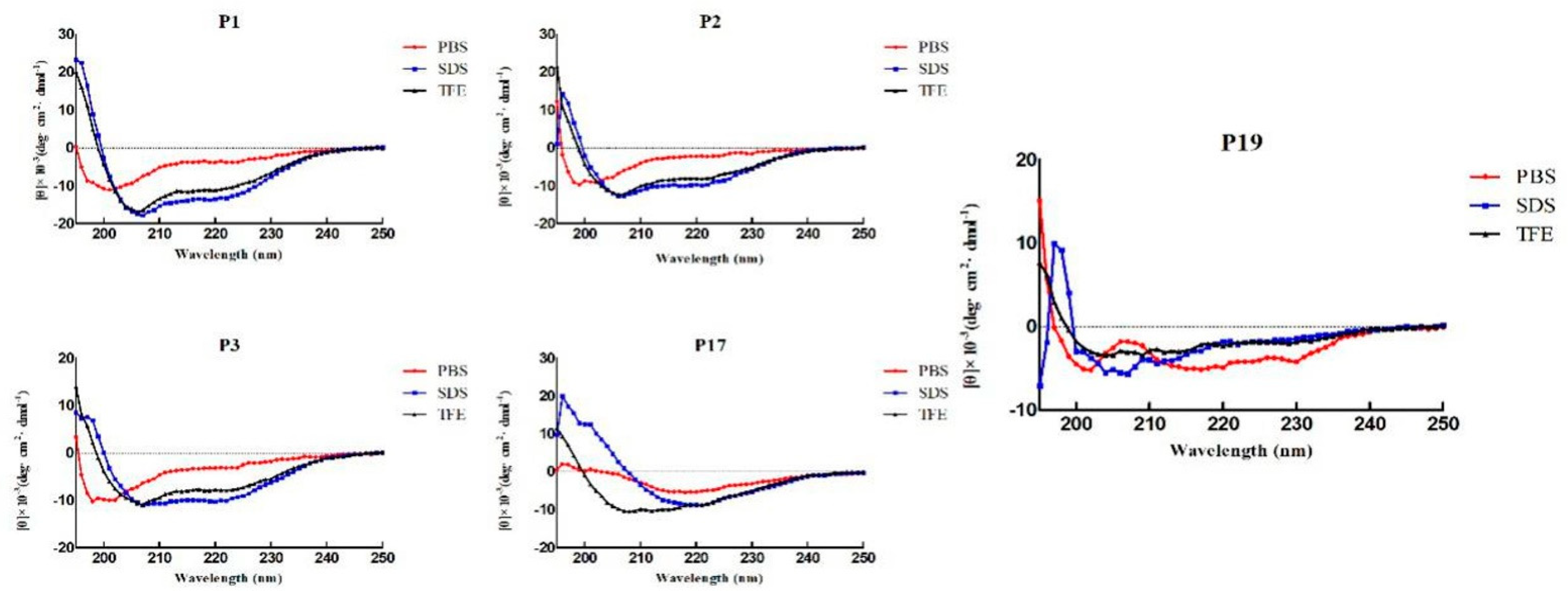





| Peptide | Sequence | Theoretical
MW | Measured
MW | Net Charge | H a | μHrel b |
|---|---|---|---|---|---|---|
| P1 | RRLALWLALRR-NH2 | 1422.79 | 1422.6 | 5 | 0.512 | 0.201 |
| P2 | RRLSLWLSLRR-NH2 | 1454.79 | 1454.6 | 5 | 0.448 | 0.262 |
| P3 | RRLCLWLCLRR-NH2 | 1486.92 | 1486.6 | 5 | 0.735 | 0.014 |
| P17 | RRISIWISIRR-NH2 | 1454.79 | 1454.8 | 5 | 0.485 | 0.267 |
| P19 | RRFSFWFSFRR-NH2 | 1590.86 | 1590.8 | 5 | 0.481 | 0.266 |
| Peptides | MICs a (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| E. coli 25922 | E. coli UB1005 | S. aureus 29213 | S. aureus 25923 | L. rhamnosus 1.0385 | L. plantarum 7469 | L. rhamnosus 1.0911 | S. thermophilus YM-C | |
| P1 | 4 | 2 | 8 | 8 | 8 | >128 | 64 | 128 |
| P2 | 4 | 2 | 4 | 16 | 8 | 128 | 8 | >128 |
| P3 | 4 | 2 | 8 | 8 | >128 | 128 | 128 | >128 |
| P17 | 128 | 16 | 128 | 128 | 8 | >128 | 32 | >128 |
| P19 | 128 | 128 | 128 | >128 | >128 | >128 | >128 | >128 |
| Melittin | 2 | 2 | 2 | 2 | >128 | >128 | >128 | >128 |
| Amphotericin B | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Fluconazole | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Peptides | MICs (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| C. albicans cgmcc 2.2086 | C. tropicalis cgmcc 2.1975 | C. parapsilosis cgmcc 2.3989 | C. albicans SP3902 | C. albicans SP3903 | C. albicans SP3937 | C. albicans 56214 | C. albicans Isolated from Alveolar Fluid | |
| P1 | 8 | 2 | 8 | 32 | 16 | 16 | 16 | 8 |
| P2 | 4 | 2 | 8 | 16 | 16 | 8 | 8 | 8 |
| P3 | 8 | 4 | 16 | 16 | 16 | 16 | 16 | 16 |
| P17 | 4 | 1 | 1 | 4 | 2 | 4 | 4 | 8 |
| P19 | 2 | 2 | 2 | 4 | 2 | 4 | 4 | 4 |
| Melittin | 4 | 2 | 2 | 4 | 2 | 4 | 2 | 4 |
| Amphotericin B | 1 | 1 | 2 | 0.5 | 0.5 | 0.5 | 1 | 1 |
| Fluconazole | 1 | 4 | 4 | 16 | 32 | 16 | >128 | 2 |
| Peptide | Control | NaCl | KCl | MgCl2 | NH4Cl | ZnCl2 | FeCl3 | pH = 6 |
|---|---|---|---|---|---|---|---|---|
| P19 | 2 | 32 | 4 | 4 | 4 | 4 | 4 | 4 |
| Melittin | 4 | 8 | 4 | 8 | 4 | 4 | 4 | 4 |
| Fluconazole | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| Amphotericin B | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, S.; Li, Q.; Wu, H.; Li, J.; Chang, Y.-F.; Shang, L.; Li, J.; Wang, Z.; Shan, A. Selective Antifungal Activity and Fungal Biofilm Inhibition of Tryptophan Center Symmetrical Short Peptide. Int. J. Mol. Sci. 2021, 22, 8231. https://doi.org/10.3390/ijms22158231
Chou S, Li Q, Wu H, Li J, Chang Y-F, Shang L, Li J, Wang Z, Shan A. Selective Antifungal Activity and Fungal Biofilm Inhibition of Tryptophan Center Symmetrical Short Peptide. International Journal of Molecular Sciences. 2021; 22(15):8231. https://doi.org/10.3390/ijms22158231
Chicago/Turabian StyleChou, Shuli, Qiuke Li, Hua Wu, Jinze Li, Yung-Fu Chang, Lu Shang, Jiawei Li, Zhihua Wang, and Anshan Shan. 2021. "Selective Antifungal Activity and Fungal Biofilm Inhibition of Tryptophan Center Symmetrical Short Peptide" International Journal of Molecular Sciences 22, no. 15: 8231. https://doi.org/10.3390/ijms22158231






