Genome-Wide Identification and Transcriptional Expression Analysis of Annexin Genes in Capsicum annuum and Characterization of CaAnn9 in Salt Tolerance
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of Ann in Pepper
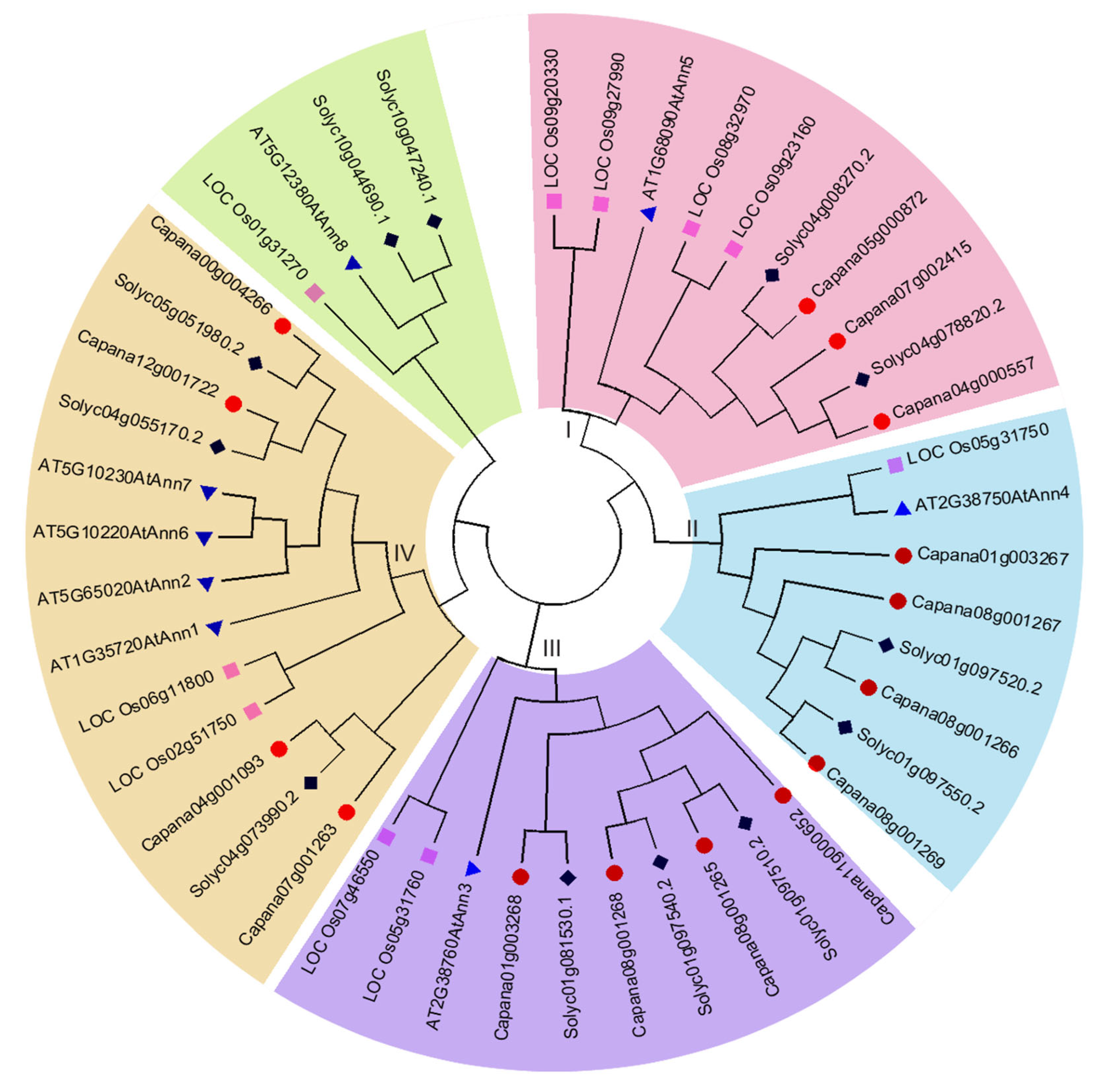
2.2. Conserved Regions and Phylogenetic Analysis of CaAnn Proteins
2.3. CaAnn Locations and Gene and Protein Structures
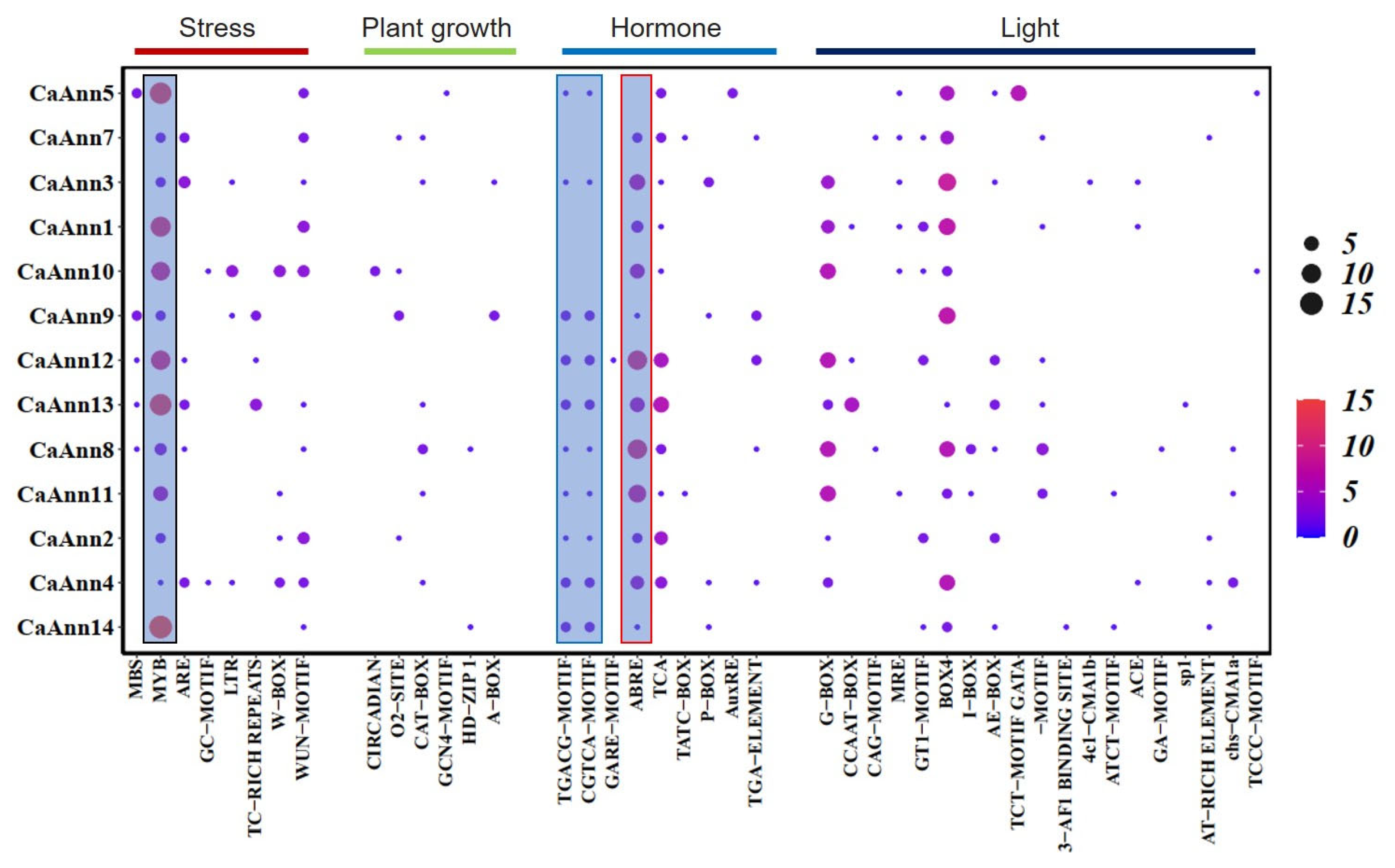
2.4. Analysis of Cis-Elements in CaAnn Gene Promoter Regions
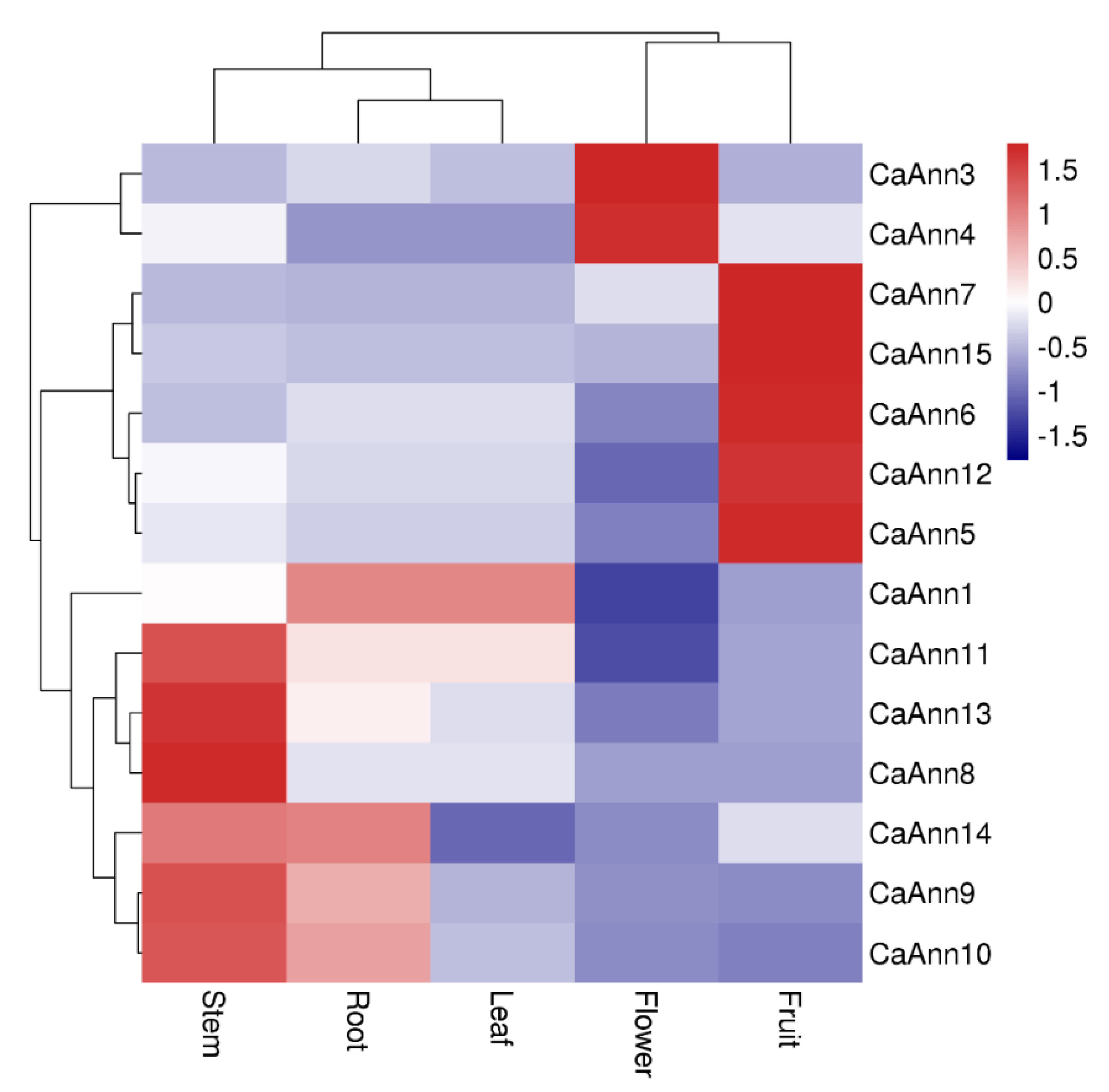
2.5. Expression Profile of CaAnn Genes in Different Tissues
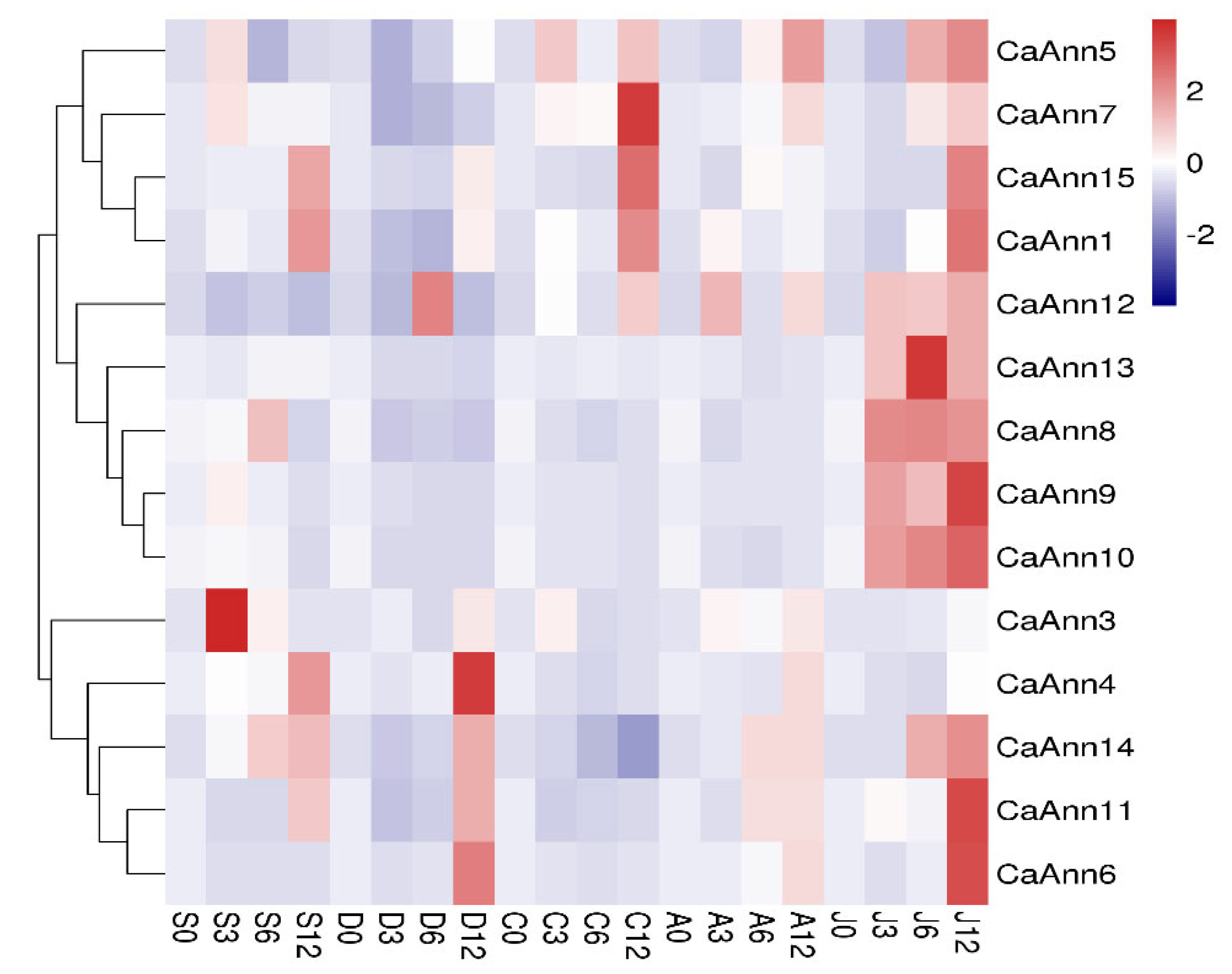
2.6. Expression Profile of CaAnn Genes in Response to Abiotic Stress and Hormonal Treatment
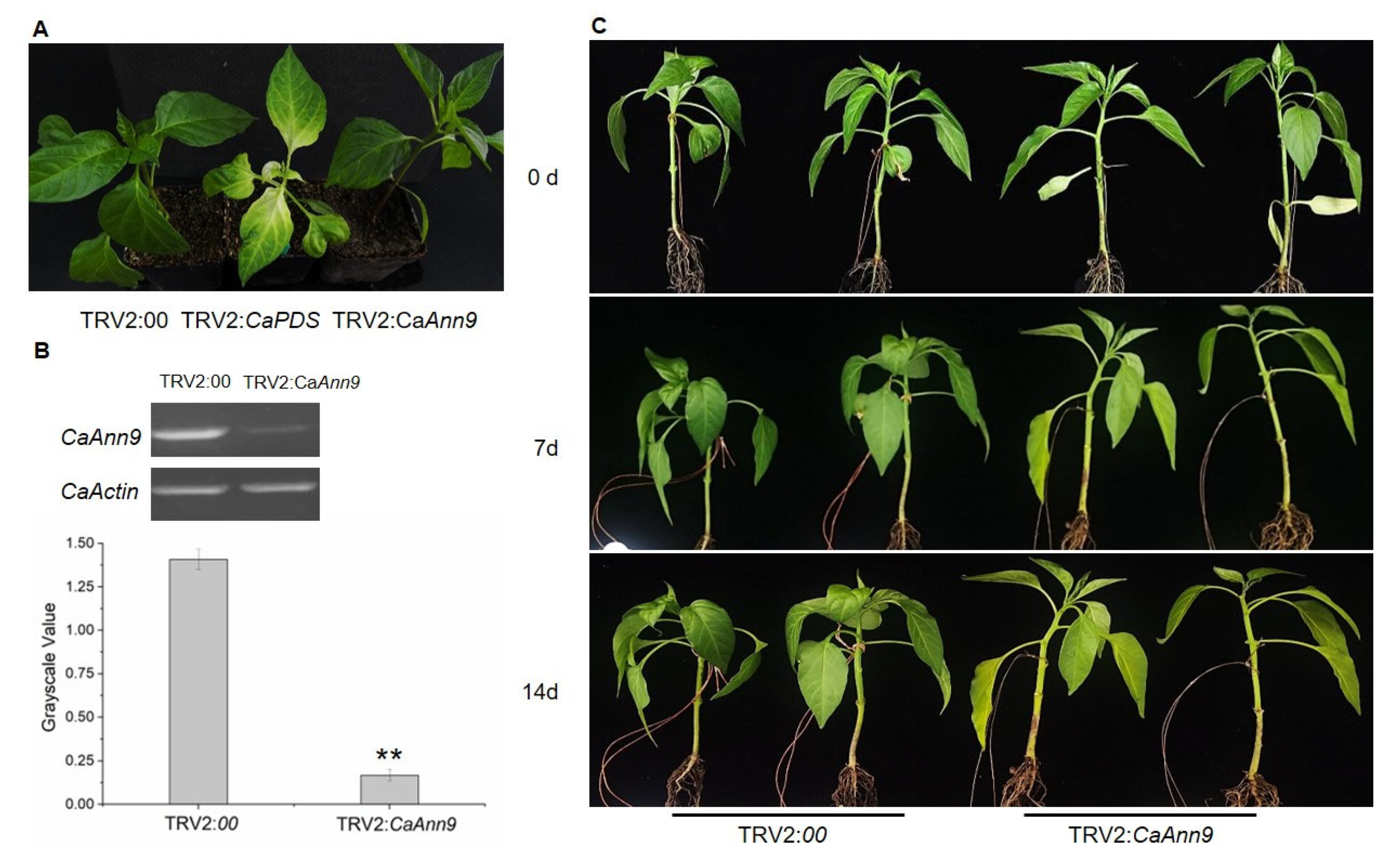
2.7. CaAnn9 Is Involved in Salt Tolerance
3. Discussion
4. Materials and Methods
4.1. Identification of Pepper Annexins
4.2. Phylogenetic Analysis
4.3. Motif Analysis of CaAnn Proteins
4.4. Plant Materials and Treatments
4.5. Expression Analysis of CaAnn Genes
4.6. Virus-Induced Gene Silencing (VIGS) of Pepper CaAnn9 Gene
4.7. Chlorophyll Content
4.8. Oxidative Damage and Antioxidant Enzyme Activities
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Laohavisit, A.; Davies, J.M. Annexins. New Phytol. 2011, 189, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, J.; Laohavisit, A.; MacPherson, N.; Webb, A.; Brownlee, C.; Battey, N.H.; Davies, J.M. Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 2008, 59, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Konopka-Postupolska, D.; Clark, G.B.; Hofmann, A. Structure, function and membrane interactions of plant annexins: An up-date. Plant Sci. 2011, 181, 230–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saad, R.B.; Ben Romdhane, W.; Ben Hsouna, A.; Mihoubi, W.; Harbaoui, M.; Brini, F. Insights into plant annexins function in abiotic and biotic stress tolerance. Plant Signal. Behav. 2020, 15, 1699264. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liao, L.; Xie, S.; Yao, M.; Xie, P.; Liu, W.; Kang, Y.; Huang, L.; Wang, M.; Qian, L.; et al. Comprehensive analyses of the annexin (ANN) gene family in Brassica rapa, Brassica oleracea and Brassica napus reveals their roles in stress response. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Boustead, C.M.; Smallwood, M.; Small, H.; Bowles, D.J.; Walker, J.H. Identification of calcium-dependent phospholipid-binding proteins in higher plant cells. FEBS Lett. 1989, 244, 456–460. [Google Scholar] [CrossRef] [Green Version]
- Clark, G.B.; Morgan, R.O.; Fernandez, M.P.; Roux, S.J. Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol. 2012, 196, 695–712. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, D.D.; Zhang, J.; Xia, H.; Wang, X.L.; Li, Y.; Li, X.B. Cotton AnnGh3 encoding an annexin protein is preferentially expressed in fibers and promotes initiation and elongation of leaf trichomes in transgenic Arabidopsis. J. Integr. Plant Biol. 2013, 55, 902–916. [Google Scholar] [CrossRef]
- Zhu, J.G.; Wu, X.R.; Yuan, S.J.; Qian, D.; Nan, Q.; An, L.Z.; Xiang, Y. Annexin5 plays a vital role in Arabidopsis pollen de-velopment via Ca2+-dependent membrane trafficking. PLoS ONE 2014, 9, e102407. [Google Scholar]
- Tang, W.; He, Y.; Tu, L.; Wang, M.; Li, Y.; Ruan, Y.-L.; Zhang, X. Down-regulating annexin gene GhAnn2 inhibits cotton fiber elongation and decreases Ca2+ influx at the cell apex. Plant Mol. Biol. 2014, 85, 613–625. [Google Scholar] [CrossRef]
- Zhang, F.; Jin, X.X.; Wang, L.K.; Li, S.F.; Wu, S.; Cheng, C.Z.; Zhang, T.Z.; Guo, W.Z. A cotton annexin affects fiber elongation and secondary cell wall biosynthesis associated with Ca2+-influx, ROS homeostasis, and actin filament reorganization. Plant Physiol. 2016, 171, 1750–1770. [Google Scholar] [CrossRef] [Green Version]
- Huh, S.M.; Noh, E.K.; Kim, H.G.; Jeon, B.W.; Bae, K.; Hu, H.-C.; Kwak, J.M.; Park, O.K. Arabidopsis annexins AnnAt1 and AnnAt4 Interact with each other and regulate drought and salt stress responses. Plant Cell Physiol. 2010, 51, 1499–1514. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ma, X.; Wang, H.; Li, B.; Clark, G.; Guo, Y.; Roux, S.; Sun, D.; Tang, W. Proteomic study of microsomal proteins reveals a key role for Arabidopsis Annexin 1 in dediating heat stress-induced increase in intracellular calcium levels. Mol. Cell. Proteom. 2015, 14, 686–694. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K.A.; Davies, J.M.; Knight, H.; Knight, M.R.; et al. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Zhang, Q.; Liu, D.; Wang, H.; Yin, J.; Wang, R.; He, M.; Cui, M.; Shang, Z.; Wang, D.; et al. A calcium-binding protein, rice annexin OsANN1, enhances heat stress tolerance by modulating the production of H2O2. J. Exp. Bot. 2015, 66, 5853–5866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.F.; Zhang, Q.; Yang, X.; Han, J.B.; Zhu, Z.G. OsANN3, a calcium-dependent lipid binding annexin is a positive regulator of ABA-dependent stress tolerance in rice. Plant Sci. 2019, 284, 212–220. [Google Scholar] [CrossRef]
- Ahmed, I.; Yadav, D.; Shukla, P.; Vineeth, T.; Sharma, P.; Kirti, P. Constitutive expression of Brassica juncea annexin, AnnBj2 confers salt tolerance and glucose and ABA insensitivity in mustard transgenic plants. Plant Sci. 2017, 265, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.L.; Li, L.J.; Liu, Q.; Liu, P.; Li, S.; Yang, D.; Chen, Y.P.; Pagnotta, S.; Favery, B.; Abad, P.; et al. A MIF-like effector sup-presses plant immunity and facilitates nematode parasitism by interacting with plant annexins. J. Exp. Bot. 2019, 70, 5943–5958. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, Y.; Lei, Y.; Li, Q.; Zhao, J.; Li, Y.; Fan, J.; Xiao, S.; Wang, W. ANNEXIN 8 negatively regulates RPW8.1-mediated cell death and disease resistance in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 378–392. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, J.; Zhang, L.; Zuo, K. A Cotton Annexin protein AnxGb6 regulates fiber elongation through its interaction with Actin 1. PLoS ONE 2013, 8, e66160. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.C.; Zheng, Y.; Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017, 216, 163–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, C.; Zhou, L.; Shan, L.B.; Li, F.J.; Li, Z.H. Phosphatase GhDsPTP3a interacts with annexin protein GhANN8b to reversely regulate salt tolerance in cotton (Gossypium spp.). New Phytol. 2019, 223, 1856–1872. [Google Scholar] [CrossRef]
- Tajima, R.; Kato, Y. Comparison of threshold algorithms for automatic image processing of rice roots using freeware ImageJ. Field Crops Res. 2011, 121, 460–463. [Google Scholar] [CrossRef]
- Cantero, A.; Barthakur, S.; Bushart, T.J.; Chou, S.; Morgan, R.O.; Fernandez, M.P.; Clark, G.B.; Roux, S.J. Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant Physiol. Biochem. 2006, 44, 13–24. [Google Scholar] [CrossRef]
- Jami, S.K.; Clark, G.B.; Ayele, B.T.; Roux, S.J.; Kirti, P.B. Identification and characterization of annexin gene family in rice. Plant Cell Rep. 2011, 31, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W.; et al. Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.M.; Wei, X.K.; Liao, W.X.; Huang, L.H.; Zhang, H.; Liang, S.C.; Peng, H. Molecular analysis of the annexin gene family in soybean. Biol. Plant. 2013, 57, 655–662. [Google Scholar] [CrossRef]
- Jami, S.K.; Clark, G.B.; Ayele, B.T.; Ashe, P.; Kirti, P.B. Genome-wide comparative analysis of annexin superfamily in plants. PLoS ONE 2012, 7, e47801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.B.; Li, X.L.; Han, M.; Wu, Z.Y. Genome-Wide Analysis and functional identification of the annexin gene family in maize (Zea mays L.). Plant Omics 2015, 8, 420–428. [Google Scholar]
- Laohavisit, A.; Shang, Z.; Rubio, L.; Cuin, T.A.; Very, A.-A.; Wang, A.; Mortimer, J.C.; Macpherson, N.; Coxon, K.M.; Battey, N.H.; et al. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+- and K+-permeable conductance in root Cells. Plant Cell 2012, 24, 1522–1533. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Song, J.; Clark, G.; Roux, S.J. ANN1 and ANN2 function in post-phloem sugar transport in root tips to affect primary root growth. Plant Physiol. 2018, 178, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Lichocka, M.; Rymaszewski, W.; Morgiewicz, K.; Barymow-Filoniuk, I.; Chlebowski, A.; Sobczak, M.; Samuel, M.A.; Schmelzer, E.; Krzymowska, M.; Hennig, J. Nucleus- and plastid-targeted annexin 5 promotes reproductive development in Arabidopsis and is essential for pollen and embryo formation. BMC Plant Biol. 2018, 18, 183. [Google Scholar] [CrossRef]
- Proust, J.; Houlne, G.; Schantz, M.L.; Schantz, R. Characterization and gene expression of an annexin during fruit development in Capsicum annuum. FEBS Lett. 1996, 383, 208–212. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.X.; Mao, L.C.; Mi, H.B.; Lu, W.J.; Ying, T.J.; Luo, Z.S. Involvement of three annexin genes in the ripening of strawberry fruit regulated by phytohormone and calcium signal transduction. Plant Cell Rep. 2016, 35, 733–743. [Google Scholar] [CrossRef]
- Jami, S.K.; Hill, R.D.; Kirti, P.B. Transcriptional regulation of annexins in Indian Mustard, Brassica juncea and detoxification of ROS in transgenic tobacco plants constitutively expressing AnnBj1. Plant Signal. Behav. 2010, 5, 618–621. [Google Scholar] [CrossRef] [Green Version]
- Jakab, G.; Ton, J.; Flors, V.; Zimmerli, L.; Métraux, J.P.; Mauch-Mani, B. Enhancing Arabidopsis salt and drought stress tol-erance by chemical priming for its abscisic acid responses. Plant Physiol. 2005, 139, 267–274. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.X.; Yan, J.X.; Wu, Y.H.; Zhang, H.B.; Mo, S.R.; Xu, X.Y.; Zhou, F.C.; Ding, H.D. Proteomic analysis by iTRAQ-PRM provides integrated insight into mechanisms of resistance in pepper to Bemisia tabaci (Gennadius). BMC Plant Biol. 2019, 19, 270. [Google Scholar] [CrossRef]
- Boyidi, P.; Shalibhadra, T.V.; Botta, H.K.; Yadav, D.; Kirti, P.B. Ectopic overexpression of abiotic stress-induced rice annexin, OsAnn5 potentiates tolerance to abiotic stresses. bioRxiv 2019. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Zhu, W.-C.; Feng, X.-H.; Jin, J.-H.; Wei, A.-M.; Gong, Z.-H. Transcription factor CaSBP12 negatively regulates aalt atress rolerance in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2020, 21, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.Y.; Zhang, J.H. Effect of abscisic acid on active oxygen species, anti-oxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 2001, 42, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.D.; He, J.; Wu, Y.; Wu, X.X.; Ge, C.L.; Wang, Y.J.; Zhong, S.L.; Peiter, E.; Liang, J.S.; Xu, W.F. The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the high-temperature stress response. Plant Physiol. 2018, 177, 633–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene ID Zunla v2.0 | Gene Name | Chr | CDS (bp) | AA | pI | Mw (kD) | Annexin Repeats | Annotation ID1 Chiltepin v2.0 | Annotation ID2 CM334 v.1.55 | Annotation ID3 CM334v.1.6 |
|---|---|---|---|---|---|---|---|---|---|---|
| Capana01g003267 | CaAnn1 | 1 | 1026 | 341 | 6.58 | 39.18 | 2 | Capang08g000204 | CA01g26520 | CAN.G1249.15 |
| Capana01g003268 | CaAnn2 | 1 | 759 | 252 | 4.99 | 28.26 | 3 | Capang08g000203 | CA01g26530 | CAN.G1249.14 |
| Capana04g000557 | CaAnn3 | 4 | 951 | 316 | 9.16 | 35.44 | 4 | Capang04g000505 | CA04g18590 | CAN.G1030.12 |
| Capana04g001093 | CaAnn4 | 4 | 945 | 314 | 5.71 | 35.85 | 4 | Capang04g001032 | CA00g85100 | CAN.G1836.3 |
| Capana05g000872 | CaAnn5 | 5 | 951 | 316 | 9.56 | 35.95 | 4 | Capang00g002303 | CAN.G966.1 | |
| Capana07g001263 | CaAnn6 | 7 | 498 | 165 | 5.96 | 19.06 | 2 | Capang07g001288 | CA07g10540 | CAN.G834.5 |
| Capana07g002415 | CaAnn7 | 7 | 951 | 316 | 7.18 | 35.66 | 4 | Capang07g002413 | CA07g20670 | CAN.G623.41 |
| Capana08g001265 | CaAnn8 | 8 | 957/975 | 318/324 | 6.47 | 35.82 | 4 | Capang01g003847 | CAN.G134.113 | |
| Capana08g001266 | CaAnn9 | 8 | 951 | 316 | 6.28 | 36.27 | 3 | Capang01g003848 | CA08g10470 | CAN.G134.114 |
| Capana08g001267 | CaAnn10 | 8 | 948 | 315 | 5.81 | 36.29 | 3 | Capang01g003849 | CA08g10480 | CAN.G134.115 |
| Capana08g001268 | CaAnn11 | 8 | 957 | 318 | 6.91 | 35.8 | 4 | Capang01g003850 | CA08g10490 | CAN.G134.116 |
| Capana08g001269 | CaAnn12 | 8 | 945 | 314 | 6.39 | 36.26 | 3 | Capang01g003851 | CAN.G134.117 | |
| Capana11g000652 | CaAnn13 | 11 | 390/459 | 129/152 | 7.11 | 40.93 | 1 | Capang11g000587 | CA11g14100 | CAN.G84.41 |
| Capana12g001722 | CaAnn14 | 12 | 951 | 316 | 5.36 | 36.14 | 4 | Capang00g001945 | CA00g83440 | CAN.G1795.1 |
| Capana00g004266 | CaAnn15 | 459 | 152 | 5.51 | 17.39 | 2 | Capang00g00115 | CA05g13700 | CAN.G151.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Ren, Y.; Jiang, H.; Wang, Y.; Yan, J.; Xu, X.; Zhou, F.; Ding, H. Genome-Wide Identification and Transcriptional Expression Analysis of Annexin Genes in Capsicum annuum and Characterization of CaAnn9 in Salt Tolerance. Int. J. Mol. Sci. 2021, 22, 8667. https://doi.org/10.3390/ijms22168667
Wu X, Ren Y, Jiang H, Wang Y, Yan J, Xu X, Zhou F, Ding H. Genome-Wide Identification and Transcriptional Expression Analysis of Annexin Genes in Capsicum annuum and Characterization of CaAnn9 in Salt Tolerance. International Journal of Molecular Sciences. 2021; 22(16):8667. https://doi.org/10.3390/ijms22168667
Chicago/Turabian StyleWu, Xiaoxia, Yan Ren, Hailong Jiang, Yan Wang, Jiaxing Yan, Xiaoying Xu, Fucai Zhou, and Haidong Ding. 2021. "Genome-Wide Identification and Transcriptional Expression Analysis of Annexin Genes in Capsicum annuum and Characterization of CaAnn9 in Salt Tolerance" International Journal of Molecular Sciences 22, no. 16: 8667. https://doi.org/10.3390/ijms22168667






