Genome-Wide Characterization and Expression Analysis of the SBP-Box Gene Family in Sweet Orange (Citrus sinensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Phylogenetic Analysis of CsSBP Genes in Sweet Orange
2.2. Chromosomal Location and Duplication Events among CsSBP Genes
2.3. Analysis of CsSBP Properties and Conserved Motifs
2.4. Plant Materials and Stress Treatments
2.5. RNA Isolation and Quantitative Real-Time PCR
3. Results and Discussion
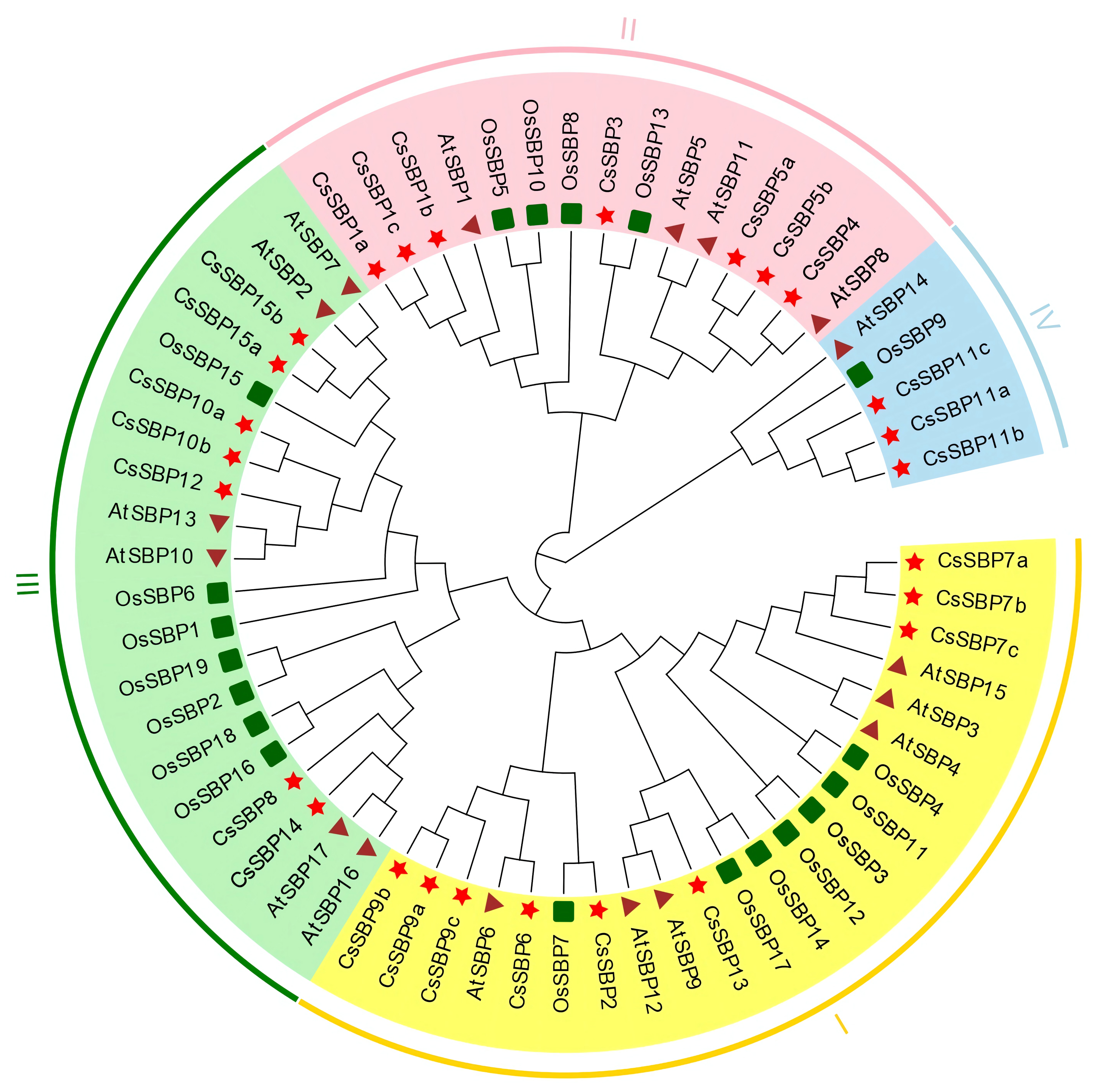
3.1. Identification and Phylogenetic Analysis of Citrus SBP Proteins
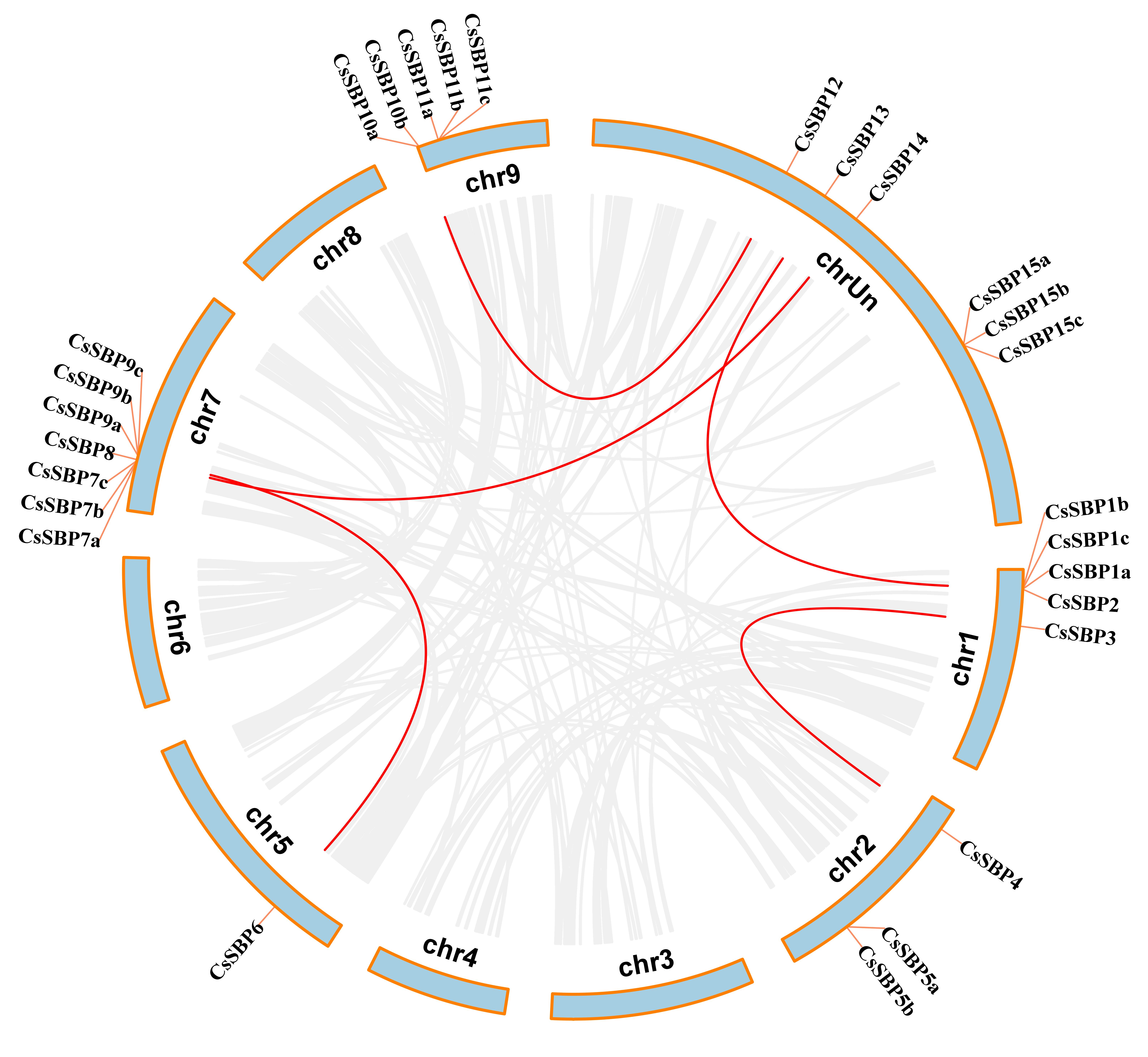
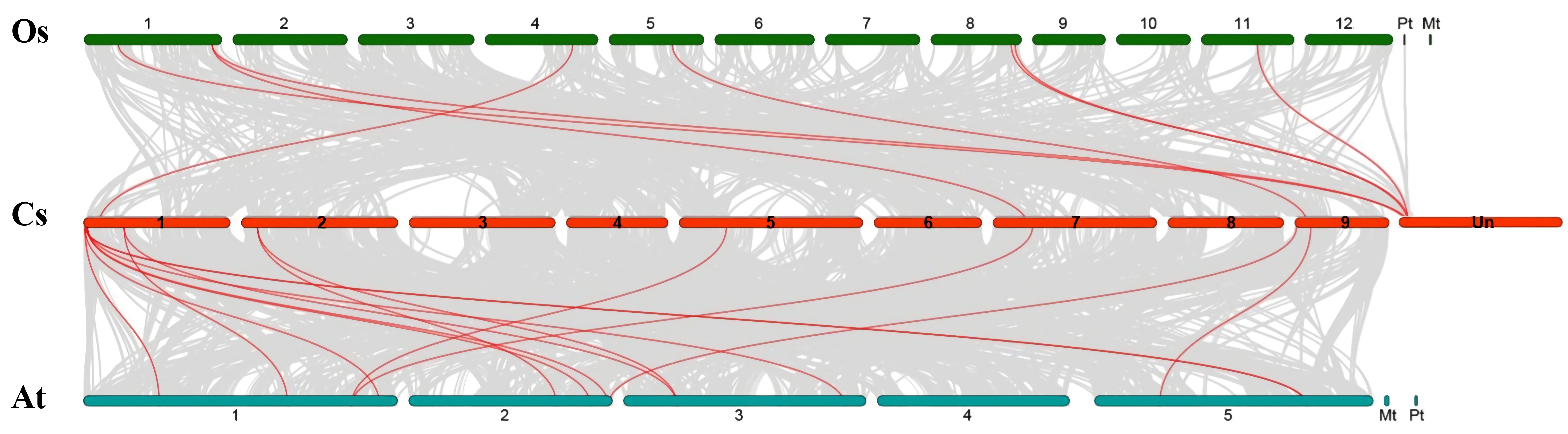
3.2. Chromosomal Locations, Evolutionary Origin, and Divergence of CsSBP Genes
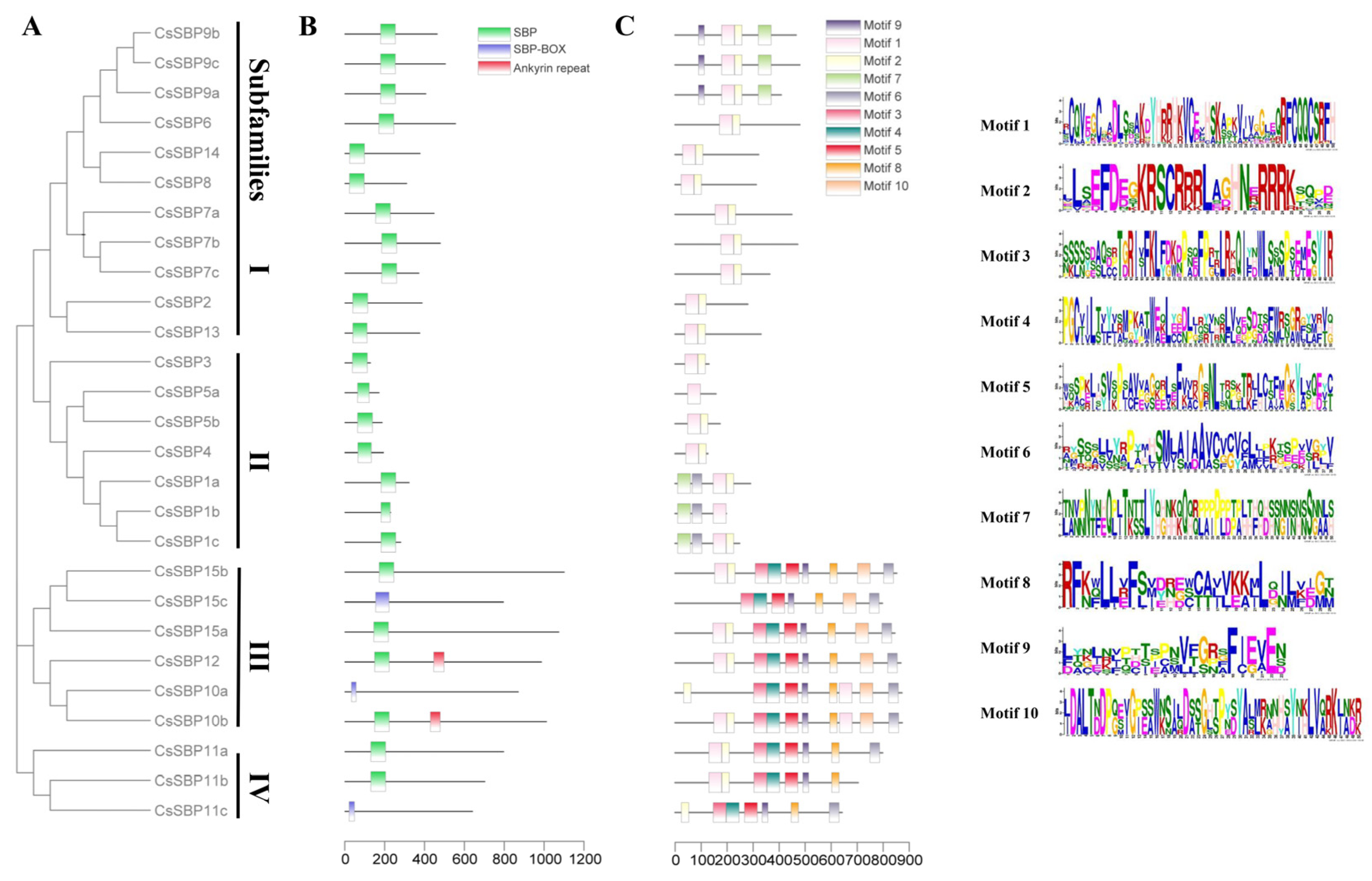
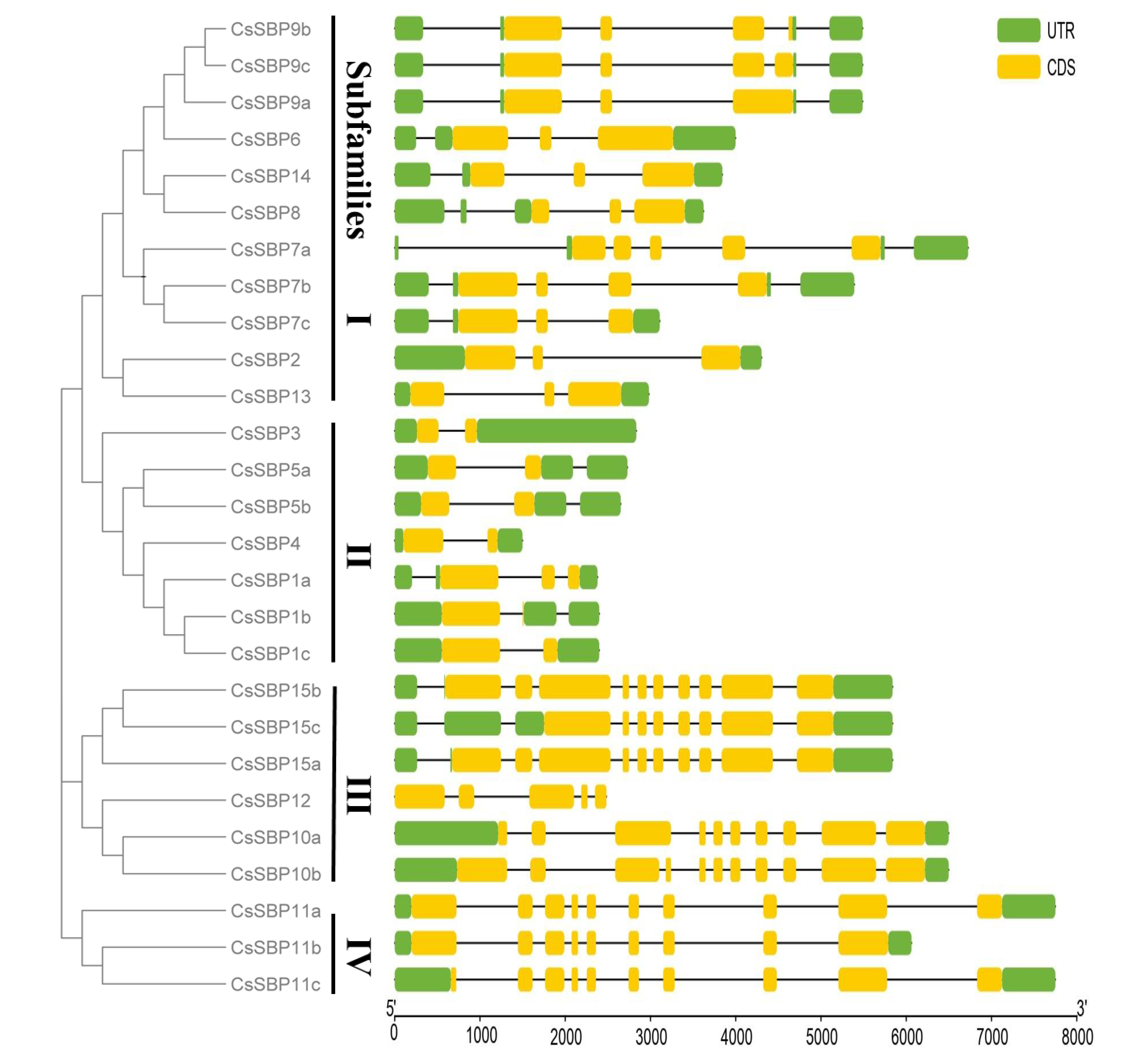
3.3. Structural Analysis of SBP Family Genes in Sweet Orange
3.4. Prediction of CRE-Involved Pathways, CsSBP Targets of miRNA
3.5. The Expression Patterns of CsSBP Genes in Various Tissues
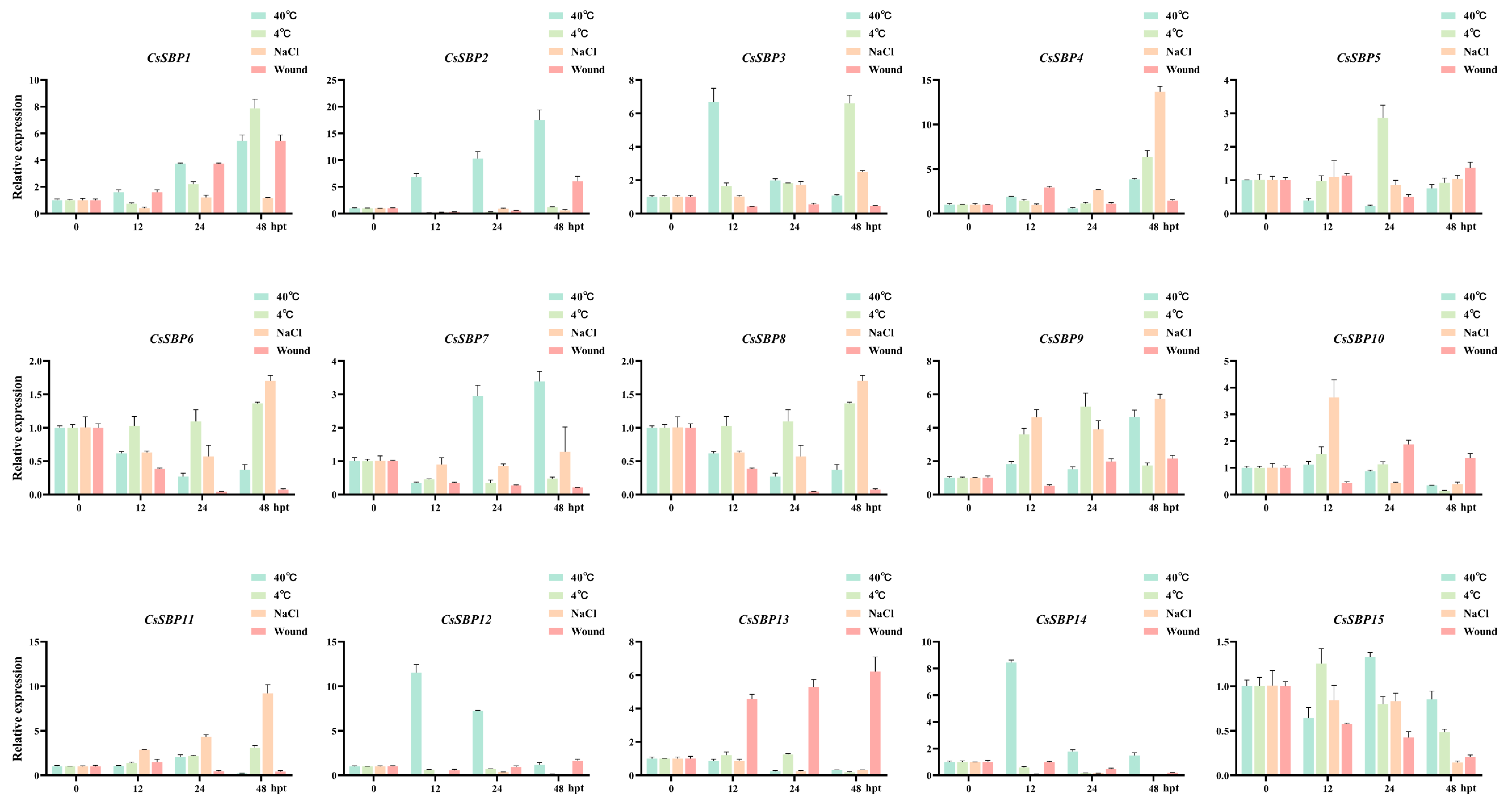
3.6. Expression Profiles of CsSBP Genes under Abiotic and Biotic Stresses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shin, Y.H.; Ko, E.J.; Kim, S.J.; Hyun, H.N.; Jeun, Y.C. Suppression of Melanose Caused by Diaporthe Citri on Citrus Leaves Pretreated with Bio-Sulfur. Plant Pathol. J. 2019, 35, 417–424. [Google Scholar] [CrossRef]
- Tan, H.-W.; Song, X.-M.; Duan, W.-K.; Wang, Y.; Hou, X.-L. Genome-Wide Analysis of the SBP-Box Gene Family in Chinese Cabbage (Brassica rapa subsp. pekinensis). Genome 2015, 58, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Li, W.; Song, N.; Tang, Z.; Liu, J.; Wang, Y.; Pan, S.; Dai, L.; Wang, B. Genome-Wide Characterization, Evolution, and Expression Profile Analysis of GATA Transcription Factors in Brachypodium Distachyon. IJMS 2021, 22, 2026. [Google Scholar] [CrossRef]
- Yu, N.; Yang, J.-C.; Yin, G.-T.; Li, R.-S.; Zou, W.-T. Genome-Wide Characterization of the SPL Gene Family Involved in the Age Development of Jatropha Curcas. BMC Genomics 2020, 21, 368. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Q.; Zhao, Y.; Li, X.; Ma, Q. Comparative Genome Analysis of the SPL Gene Family Reveals Novel Evolutionary Features in Maize. Genet. Mol. Biol. 2019, 42, 380–394. [Google Scholar] [CrossRef]
- Song, A. Transcriptome-Wide Identification and Expression Analysis of Chrysanthemum SBP-like Transcription Factors. Plant Physiol. Biochem. 2016, 102, 10–16. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.-M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-Box Gene That Affects Pollen Sac Development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Poethig, R.S. Temporal Regulation of Shoot Development in Arabidopsis thaliana by miR156 and Its Target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual Effects of MiR156-Targeted SPL Genes and CYP78A5/KLUH on Plastochron Length and Organ Size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-H.; Ju, Y.; Seo, P.J.; Lee, J.-H.; Park, C.-M. The SOC1-SPL Module Integrates Photoperiod and Gibberellic Acid Signals to Control Flowering Time in Arabidopsis: SOC1-SPL Module in Flowering Promotion. Plant J. 2012, 69, 577–588. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of MiR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis Thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-MiR164a Targets OsNAC60 and Negatively Regulates Rice Immunity against the Blast Fungus Magnaporthe oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Wei, M.; Li, Y.; Tao, H.; Wu, H.; Chen, Z.; Li, C.; Xu, J.-H. MiR529a Controls Plant Height, Tiller Number, Panicle Architecture and Grain Size by Regulating SPL Target Genes in Rice (Oryza sativa L.). Plant Sci. 2021, 302, 110728. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Chen, P.; Zhang, X.; Liu, Q.; Li, H. Comparative Analysis of the SPL Gene Family in Five Rosaceae Species: Fragaria vesca, Malus domestica, Prunus persica, Rubus occidentalis, and Pyrus pyrifolia. Open Life Sci. 2021, 16, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Han, J.; Yang, Z.; Liu, Q.; Shuai, Y.; Xu, X.; Nie, G.; Huang, L.; Liu, W.; Zhang, X. Genome-Wide Identification, Phylogenetic Analysis, and Expression Analysis of the SPL Gene Family in Orchardgrass (Dactylis glomerata L.). Genomics 2021, 113, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Hanly, A.; Karagiannis, J.; Lu, Q.S.M.; Tian, L.; Hannoufa, A. Characterization of the Role of SPL9 in Drought Stress Tolerance in Medicago Sativa. IJMS 2020, 21, 6003. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Feng, X.-H.; Ali, M.; Jin, J.-H.; Wei, A.-M.; Khattak, A.M.; Gong, Z.-H. Identification of Pepper CaSBP08 Gene in Defense Response Against Phytophthora Capsici Infection. Front. Plant Sci. 2020, 11, 183. [Google Scholar] [CrossRef]
- Liu, M.; Shi, Z.; Zhang, X.; Wang, M.; Zhang, L.; Zheng, K.; Liu, J.; Hu, X.; Di, C.; Qian, Q.; et al. Inducible Overexpression of Ideal Plant Architecture1 Improves Both Yield and Disease Resistance in Rice. Nat. Plants 2019, 5, 389–400. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative Study of SBP-Box Gene Family in Arabidopsis and Rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Balfagón, D.; Zandalinas, S.I.; Gómez-Cadenas, A. High Temperatures Change the Perspective: Integrating Hormonal Responses in Citrus Plants under Co-Occurring Abiotic Stress Conditions. Physiol. Plant. 2019, 165, 183–197. [Google Scholar] [CrossRef]
- Hou, X.-J.; Li, S.-B.; Liu, S.-R.; Hu, C.-G.; Zhang, J.-Z. Genome-Wide Classification and Evolutionary and Expression Analyses of Citrus MYB Transcription Factor Families in Sweet Orange. PLoS ONE 2014, 9, e112375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-D.; Ling, L.-Z. Genome-Wide Identification and Evolutionary Analysis of the SBP-Box Gene Family in Castor Bean. PLoS ONE 2014, 9, e86688. [Google Scholar] [CrossRef]
- Li, J.; Gao, X.; Sang, S.; Liu, C. Genome-Wide Identification, Phylogeny, and Expression Analysis of the SBP-Box Gene Family in Euphorbiaceae. BMC Genomics 2019, 20, 912. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Jin, J.-H.; He, Y.-M.; Lu, B.-Y.; Li, D.-W.; Chai, W.-G.; Khan, A.; Gong, Z.-H. Genome-Wide Identification and Analysis of the SBP-Box Family Genes under Phytophthora Capsici Stress in Pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic Organization, Phylogenetic Comparison and Differential Expression of the SBP-Box Family of Transcription Factors in Tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Q.; Zhang, Y.; Li, Z.; Guo, J.; Chen, X.; Zhang, G. Genome-Wide Identification, Characterization, and Expression Patterns Analysis of the SBP-Box Gene Family in Wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 17250. [Google Scholar] [CrossRef]
- Pan, F.; Wang, Y.; Liu, H.; Wu, M.; Chu, W.; Chen, D.; Xiang, Y. Genome-Wide Identification and Expression Analysis of SBP-like Transcription Factor Genes in Moso Bamboo (Phyllostachys edulis). BMC Genomics 2017, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Zhou, H.; Sheng, S.; Cao, M.; Li, Y.; Pang, X. Genome-Wide Organization and Expression Profiling of the SBP-Box Gene Family in Chinese Jujube (Ziziphus jujuba Mill.). IJMS 2017, 18, 1734. [Google Scholar] [CrossRef] [Green Version]
- Araki, R.; Mermod, M.; Yamasaki, H.; Kamiya, T.; Fujiwara, T.; Shikanai, T. SPL7 Locally Regulates Copper-Homeostasis-Related Genes in Arabidopsis. J. Plant Physiol. 2018, 224–225, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Noh, H.; Mun, J.; Gu, C.; Sever, S.; Park, S. Anks1a Regulates COPII-Mediated Anterograde Transport of Receptor Tyrosine Kinases Critical for Tumorigenesis. Nat. Commun. 2016, 7, 12799. [Google Scholar] [CrossRef] [Green Version]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a Plant-Specific SBP-Domain Transcription Factor, Participates in Plant Development and Sensitivity to Fumonisin B1: AtSPL14 Functions in Plant Development. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deb, A.; Grewal, R.K.; Kundu, S. Regulatory Cross-Talks and Cascades in Rice Hormone Biosynthesis Pathways Contribute to Stress Signaling. Front. Plant Sci. 2016, 7, 1303. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Gao, X.; Zhang, X.; Liu, C. Dynamic Expansion and Functional Evolutionary Profiles of Plant Conservative Gene Family SBP-Box in Twenty Two Flowering Plants and the Origin of MiR156. Biomolecules 2020, 10, 757. [Google Scholar] [CrossRef]
- Liang, C.; Liu, H.; Hao, J.; Li, J.; Luo, L. Expression Profiling and Regulatory Network of Cucumber MicroRNAs and Their Putative Target Genes in Response to Cucumber Green Mottle Mosaic Virus Infection. Arch. Virol. 2019, 164, 1121–1134. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P.; et al. The Draft Genome of Sweet Orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Gai, W.-X.; Ma, X.; Qiao, Y.-M.; Shi, B.-H.; ul Haq, S.; Li, Q.-H.; Wei, A.-M.; Liu, K.-K.; Gong, Z.-H. Characterization of the BZIP Transcription Factor Family in Pepper (Capsicum annuum L.): CabZIP25 Positively Modulates the Salt Tolerance. Front. Plant Sci. 2020, 11, 139. [Google Scholar] [CrossRef]
- Hong, Y.; Ahmad, N.; Tian, Y.; Liu, J.; Wang, L.; Wang, G.; Liu, X.; Dong, Y.; Wang, F.; Liu, W.; et al. Genome-Wide Identification, Expression Analysis, and Subcellular Localization of Carthamus Tinctorius BHLH Transcription Factors. IJMS 2019, 20, 3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, K.; Tan, P.; Guo, W.; Yue, Y.; Fan, X.; Wu, J. Heterologous Expression of a Novel Zoysia Japonica C2H2 Zinc Finger Gene, ZjZFN1, Improved Salt Tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 1159. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The MiR156/SPL Module Regulates Apple Salt Stress Tolerance by Activating MdWRKY100 Expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.; Arshad, M.; Hannoufa, A. Alfalfa Response to Heat Stress Is Modulated by MicroRNA156. Physiol. Plant. 2019, 165, 830–842. [Google Scholar] [CrossRef]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.-M.; Thiruvengadam, M. Characterizing the Role of the MiR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-X.; Ali, M.; Feng, X.-H.; Jin, J.-H.; Huang, L.-J.; Khan, A.; Lv, J.-G.; Gao, S.-Y.; Luo, D.-X.; Gong, Z.-H. A Novel Transcription Factor CaSBP12 Gene Negatively Regulates the Defense Response against Phytophthora Capsici in Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2019, 20, 48. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-X.; Feng, X.-H.; Jin, J.-H.; Khan, A.; Guo, W.-L.; Du, X.-H.; Gong, Z.-H. CaSBP11 Participates in the Defense Response of Pepper to Phytophthora Capsici through Regulating the Expression of Defense-Related Genes. IJMS 2020, 21, 9065. [Google Scholar] [CrossRef]
- Mao, Y.-B.; Liu, Y.-Q.; Chen, D.-Y.; Chen, F.-Y.; Fang, X.; Hong, G.-J.; Wang, L.-J.; Wang, J.-W.; Chen, X.-Y. Jasmonate Response Decay and Defense Metabolite Accumulation Contributes to Age-Regulated Dynamics of Plant Insect Resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Hong, G.; Li, L.; Zhang, X.; Kong, Y.; Sun, Z.; Li, J.; Chen, J.; He, Y. MiR156/SPL9 Regulates Reactive Oxygen Species Accumulation and Immune Response in Arabidopsis thaliana. Phytopathology 2019, 109, 632–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, N.; Cheng, Y.; Peng, W.; Peng, E.; Zhao, Z.; Liu, T.; Yi, T.; Dai, L.; Wang, B.; Hong, Y. Genome-Wide Characterization and Expression Analysis of the SBP-Box Gene Family in Sweet Orange (Citrus sinensis). Int. J. Mol. Sci. 2021, 22, 8918. https://doi.org/10.3390/ijms22168918
Song N, Cheng Y, Peng W, Peng E, Zhao Z, Liu T, Yi T, Dai L, Wang B, Hong Y. Genome-Wide Characterization and Expression Analysis of the SBP-Box Gene Family in Sweet Orange (Citrus sinensis). International Journal of Molecular Sciences. 2021; 22(16):8918. https://doi.org/10.3390/ijms22168918
Chicago/Turabian StyleSong, Na, Yulin Cheng, Weiye Peng, ErPing Peng, Zengling Zhao, Tiantian Liu, Tuyong Yi, Liangying Dai, Bing Wang, and Yanyun Hong. 2021. "Genome-Wide Characterization and Expression Analysis of the SBP-Box Gene Family in Sweet Orange (Citrus sinensis)" International Journal of Molecular Sciences 22, no. 16: 8918. https://doi.org/10.3390/ijms22168918





