The First Cytoplasmic Loop in the Core Structure of the ABCC1 (Multidrug Resistance Protein 1; MRP1) Transporter Contains Multiple Amino Acids Essential for Its Expression
Abstract
:1. Introduction
2. Results
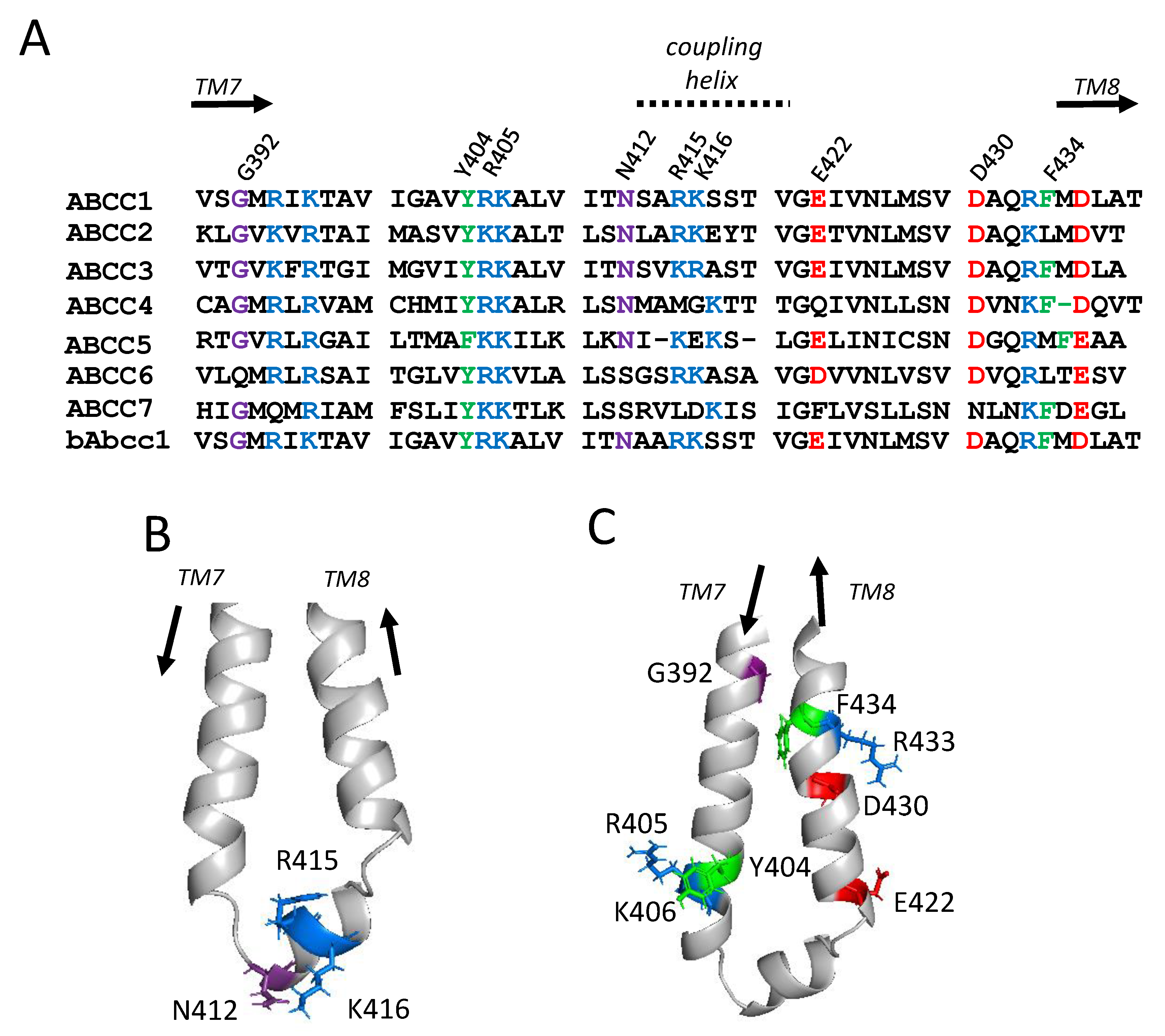
2.1. Sequence Alignments and Initial Selection of CL4 Amino Acids for Mutagenesis
2.2. Effect of Mutating Asn412, Arg415, and Lys416 in the Coupling Helix of CL4
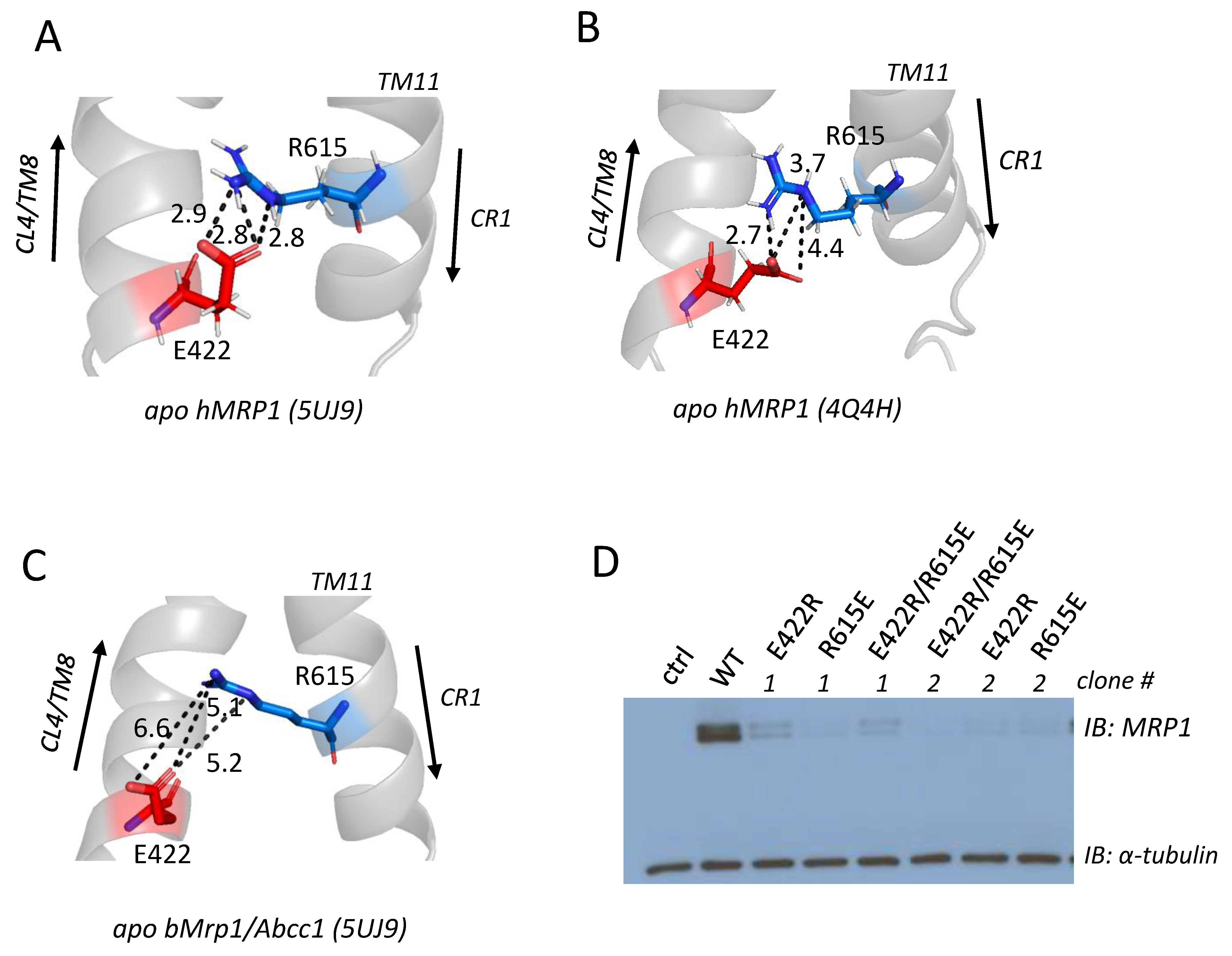
2.3. Effect of Mutating Gly392, Tyr404, and Arg405 Upstream and Glu422 and Phe434 Downstream of the CL4 Coupling Helix on hMRP1
2.4. Effect of Exchange Mutations of CL4-Glu422 and CR1-Arg615 on hMRP1 Levels in HEK Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Site-Directed Mutagenesis
4.3. Cell Culture, Transfections, and Preparation of Whole Cell Extracts and Membrane Vesicles
4.4. Immunoblotting
4.5. Vesicular Transport Assays
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Slot, A.J.; Molinski, S.V.; Cole, S.P.C. Mammalian multidrug resistance proteins (MRPs). Essays Biochem. 2011, 50, 179–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keppler, D. Multidrug resistance proteins (MRfPs, ABCCs): Importance for pathophysiology and drug therapy. Handb. Exp. Pharmacol. 2011, 201, 299–323. [Google Scholar] [CrossRef]
- Cole, S.P.C.; Bhardwaj, G.; Gerlach, J.H.; Mackie, J.E.; Grant, C.E.; Almquist, K.C.; Stewart, A.J.; Kurz, E.U.; Duncan, A.M.V.; Deeley, R.G. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 1992, 258, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.P.C. Targeting the multidrug resistance protein (MRP1, ABCC1): Past, present and future. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 95–117. [Google Scholar] [CrossRef]
- Cole, S.P.C. Multidrug resistance protein 1 (MRP1, ABCC1): A “multitasking” ABC transporter. J. Biol. Chem. 2014, 289, 30880–30888. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.P.C.; Sparks, K.E.; Fraser, K.; Loe, D.W.; Grant, C.E.; Wilson, G.M.; Deeley, R.G. Pharmacological characterization of multidrug resistant MRP-transfected human tumor cells. Cancer Res. 1994, 54, 5902–5910. [Google Scholar] [PubMed]
- Loe, D.W.; Almquist, K.C.; Deeley, R.G.; Cole, S.P.C. Multidrug resistance protein (MRP)-mediated transport of leukotriene C4 and chemotherapeutic agents in membrane vesicles: Demonstration of glutathione-dependent vincristine transport. J. Biol. Chem. 1996, 271, 9675–9682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Y.M.; Song, W.C.; Cui, H.; Cole, S.P.C.; Deeley, R.G. Glutathione stimulates sulfated estrogen transport by multidrug resistance protein 1. J. Biol. Chem. 2001, 276, 6404–6411. [Google Scholar] [CrossRef] [Green Version]
- Rothnie, A.; Callaghan, R.; Deeley, R.G.; Cole, S.P.C. Role of GSH in estrone sulfate binding and translocation by the multidrug resistance protein 1 (MRP1/ABCC1). J. Biol. Chem. 2006, 281, 13906–13914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Cui, L.; Riordan, J.R.; Chang, X. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J. Biol. Chem. 2000, 275, 20280–20287. [Google Scholar] [CrossRef] [Green Version]
- Stockner, T.; Gradisch, R.; Schmitt, L. The role of the degenerate nucleotide binding site in type I ABC exporters. FEBS Lett. 2020, 594, 3815–3838. [Google Scholar] [CrossRef]
- Rudashevskaya, E.L.; Stockner, T.; Trauner, M.; Freissmuth, M.; Chiba, P. Pharmacological correction of misfolding of ABC proteins. Drug Discov. Today Technol. 2014, 12, e87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locher, K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016, 23, 487–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conseil, G.; Arama-Chayoth, M.; Tsfadia, Y.; Cole, S.P.C. Structure-guided probing of the LTC4 binding site in human multidrug resistance protein 1 (MRP1; ABCC1). FASEB J. 2019, 33, 10692–10704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iram, S.H.; Cole, S.P.C. Expression and function of human MRP1 (ABCC1) is dependent on amino acids in cytoplasmic loop 5 and its interface with nucleotide binding domain 2. J. Biol. Chem. 2011, 286, 7202–7213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conseil, G.; Rothnie, A.; Deeley, R.G.; Cole, S.P.C. Multiple roles of charged amino acids in cytoplasmic loop 7 for expression and function of the multidrug and organic anion transporter MRP1 (ABCC1). Mol. Pharmacol. 2009, 75, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Hou, Y.X.; Campbell, C.A.; Palaniyandi, K.; Zhao, Q.; Bordner, A.J.; Chang, X.B. Glutamine residues in Q-loops of multidrug resistance protein MRP1 contribute to ATP binding via interaction with metal cofactor. Biochim. Biophys. Acta 2011, 1808, 1790–1796. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Scavetta, R.; Chang, X.B. The hydroxyl group of S685 in Walker A motif and the carboxyl group of D792 in Walker B motif of NBD1 play a crucial role for multidrug resistance protein folding and function. Biochim. Biophys. Acta 2008, 1778, 454–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, E.E.; Conseil, G.; Cole, S.P.C. Conservative amino acids in the region connecting membrane spanning domain 1 to nucleotide binding domain 1 are essential for expression of the MRP1 (ABCC1) transporter. PLoS ONE 2021, 16, e0246727. [Google Scholar] [CrossRef]
- Johnson, Z.L.; Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 2017, 168, 1075–1085. [Google Scholar] [CrossRef] [Green Version]
- Weigl, K.E.; Conseil, G.; Rothnie, A.J.; Arama, M.; Tsfadia, Y.; Cole, S.P.C. An outward-facing aromatic amino acid is crucial for signaling between the membrane-spanning and nucleotide-binding domains of multidrug resistance protein 1 (MRP1; ABCC1). Mol. Pharmacol. 2018, 94, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Conrad, S.; Kauffmann, H.-M.; Ito, K.-i.; Leslie, E.M.; Deeley, R.G.; Schrenk, D.; Cole, S.P.C. A naturally occurring mutation in MRP1 results in a selective decrease in organic anion transport and in increased doxorubicin resistance. Pharmacogenetics 2002, 12, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Haimeur, A.; Conseil, G.; Deeley, R.G.; Cole, S.P.C. Mutations of charged amino acids in or near the transmembrane helices of the second membrane spanning domain differentially affect the substrate specificity and transport activity of the multidrug resistance protein MRP1 (ABCC1). Mol. Pharmacol. 2004, 65, 1375–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, Z.L.; Chen, J. ATP binding enables substrate release from multidrug resistance protein 1. Cell 2018, 172, 81–89. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef]
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. Chem. Bio. Chem. 2002, 3, 604–617. [Google Scholar] [CrossRef]
- Loo, T.W.; Clarke, D.M. A salt bridge in intracellular loop 2 is essential for folding of human P-glycoprotein. Biochemistry 2013, 52, 3194–3196. [Google Scholar] [CrossRef] [PubMed]
- Koban, F.; El-Kasaby, A.; Häusler, C.; Stockner, T.; Simbrunner, B.M.; Sitte, H.H.; Freissmuth, M.; Sucic, S. A salt bridge linking the first intracellular loop with the C terminus facilitates the folding of the serotonin transporter. J. Biol. Chem. 2015, 290, 13263–13278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graab, P.; Bock, C.; Weiss, K.; Hirth, A.; Koller, N.; Braner, M.; Jung, J.; Loehr, F.; Tampé, R.; Behrends, C.; et al. Lysosomal targeting of the ABC transporter TAPL is determined by membrane-localized charged residues. J. Biol. Chem. 2019, 294, 7308–7323. [Google Scholar] [CrossRef]
- Ren, X.Q.; Furukawa, T.; Yamamoto, M.; Aoki, S.; Kobayashi, M.; Nakagawa, M.; Akiyama, S.I. A functional role of intracellular loops of human multidrug resistance protein 1. J. Biochem. 2006, 140, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Mercier, B.; Verlingue, C.; Lissens, W.; Silber, S.J.; Novelli, G.; Bonduelle, M.; Audrezet, M.P.; Ferec, C. Is congenital bilateral absence of vas deterens a primary form of cystic fibrosis? Analyses of the CFTR gene in 67 patients. Am. J. Hum. Genet. 1995, 56, 272–277. [Google Scholar] [CrossRef]
- Seibert, F.S.; Jia, Y.; Mathews, C.J.; Hanrahan, J.W.; Riordan, J.R.; Loo, T.W.; Clarke, D.M. Disease-associated mutations in cytoplamic loops 1 and 2 of cystic fibrosis transmembrane conductance regulator impede processing or opening of the channel. Biochemistry 1997, 36, 11966–11974. [Google Scholar] [CrossRef]
- Loo, T.W.; Clarke, D.M. Corrector VX-809 promotes interaction between cytoplasmic loop 1 and the first nucleotide-binding domain of CFTR. Biochem. Pharmacol. 2017, 136, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, A.; Chung, W.J.; Pyle, L.C.; Wang, W.; Nowotarski, K.; Mulvihill, C.M.; Ramjeesingh, M.; Hong, J.; Velu, S.E.; Lewis, H.A.; et al. Channel gating regulation by the cystic fibrosis transmembrane conductance regulator (CFTR) first cytosolic loop. J. Biol. Chem. 2016, 291, 1854–1865. [Google Scholar] [CrossRef] [Green Version]
- Vanakker, O.M.; Leroy, B.P.; Coucke, P.; Bercovitch, L.G.; Uitto, J.; Viljoen, D.; Terry, S.F.; Van Acker, P.; Matthys, D.; Loeys, B.; et al. Novel clinico-molecular insights in Pseudoxamnthoma elasticum provide an efficient molecular screening method and a comprehensive diagnostic flowchart. Hum. Mut. 2008, 29, 205. [Google Scholar] [CrossRef]
- Issa, P.C.; Tysoe, C.; Caswell, R. Late-onset pseudoxanthoma elasticum associated with a hypomorphic ABCC6 variant. Am. J. Ophthalmol. 2020, 218, 255–260. [Google Scholar] [CrossRef]
- Dougherty, D.A. The cation-π interaction. Chem. Res. 2013, 46, 885–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craven, T.W.; Cho, M.K.; Traaseth, N.J.; Bonneau, R.; Kirshenbaum, K. A miniature protein stabilized by a cation-π interaction network core. J. Am. Chem. Soc. 2016, 138, 1543–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.Y.; Grove, D.E.; de la Rosa, O.; Houck, S.A.; Sopha, P.; van Goor, F.; Hoffman, B.J.; Cyr, D.M. VX-809 corrects folding defects in cystic fibrosis transmembrane conductance regulator protein through action on membrane-spanning domain 1. Mol. Biol. Cell 2013, 24, 3016–3024. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.R. CFTR function and prospects for therapy. Annu. Rev. Biochem. 2008, 77, 701–726. [Google Scholar] [CrossRef]
- Strom, C.M.; Huang, D.; Chen, C.; Buller, A.; Peng, M.; Quan, F.; Redman, J.; Sun, W. Extensive sequencing of the cystic fibrosis transmembrane regulator gene: Assay validation and unexpected benefits of developing a comprehensive test. Genet. Med. 2003, 5, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosshard, H.R.; Marti, D.N.; Jelesarov, I. Protein stabilization by salt bridges: Concepts, experimental approaches and clarification of some misunderstandings. J. Mol. Recognit. 2004, 17, 1–16. [Google Scholar] [CrossRef]
- Hipfner, D.R.; Almquist, K.C.; Stride, B.D.; Deeley, R.G.; Cole, S.P.C. Location of a protease-hypersensitive region in the multidrug resistance protein (MRP) by mapping of the epitope of MRP-specific monoclonal antibody QCRL-1. Cancer Res. 1996, 56, 3307–3314. [Google Scholar] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Shaw, V.S.; Mohammadi, M.; Quinn, J.A.; Vashisth, H.; Neubig, R.R. An interhelical salt bridge controls flexibility and inhibitor potency for regulators of G-protein signaling proteins 4, 8, and 19. Mol. Pharmacol. 2019, 96, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Bin Kanner, Y.; Ganoth, A.; Tsfadia, Y. Extracellular mutation induces an allosteric effect across the membrane and hampers the activity of MRP1 (ABCC1). Sci. Rep. 2021, 11, 12024. [Google Scholar] [CrossRef]
- Lewinson, O.; Orelle, C.; Seeger, M.A. Structures of ABC transporters: Handle with care. FEBS Lett. 2020, 594, 3799–3814. [Google Scholar] [CrossRef] [PubMed]



| Mutant | hMRP1 Levels (% WT-hMRP1) (n) | |
|---|---|---|
| WCE | MV | |
| N412A | 133 ± 15 (3) | 119 ± 28 (3) |
| R415A | 83 ± 16 (5) | 81 ± 31 (3) |
| R415K | 104 ± 21 (3) | 85 ± 26 (4) |
| K416A | 103 ± 21 (3) | 97 ± 39 (3) |
| K416R | 138 ± 12 (3) | 127 ± 20 (3) |
| Mutant. | Transport Activity (% WT-hMRP1) | ||
|---|---|---|---|
| LTC4 | E217βG | E13SO4 (+ S-Me GSH) | |
| HEK ctrl | 6 ± 4 * | 0 ± 0 * | 7 ± 2 * |
| WT-hMRP1 | 100 ± 10 | 100 ± 10 | 100 ± 15 |
| Y404F | 90 ± 2 | 102 ± 9 | 107 ± 17 |
| E422D | 84 ± 16 | 109 ± 9 | 89 ± 14 |
| D430E | 87 ± 11 | 102 ± 4 | 104 ± 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conseil, G.; Cole, S.P.C. The First Cytoplasmic Loop in the Core Structure of the ABCC1 (Multidrug Resistance Protein 1; MRP1) Transporter Contains Multiple Amino Acids Essential for Its Expression. Int. J. Mol. Sci. 2021, 22, 9710. https://doi.org/10.3390/ijms22189710
Conseil G, Cole SPC. The First Cytoplasmic Loop in the Core Structure of the ABCC1 (Multidrug Resistance Protein 1; MRP1) Transporter Contains Multiple Amino Acids Essential for Its Expression. International Journal of Molecular Sciences. 2021; 22(18):9710. https://doi.org/10.3390/ijms22189710
Chicago/Turabian StyleConseil, Gwenaëlle, and Susan P. C. Cole. 2021. "The First Cytoplasmic Loop in the Core Structure of the ABCC1 (Multidrug Resistance Protein 1; MRP1) Transporter Contains Multiple Amino Acids Essential for Its Expression" International Journal of Molecular Sciences 22, no. 18: 9710. https://doi.org/10.3390/ijms22189710





