Photoactivatable nanoCRISPR/Cas9 System Based on crRNA Reversibly Immobilized on Carbon Nanoparticles
Abstract
:1. Introduction
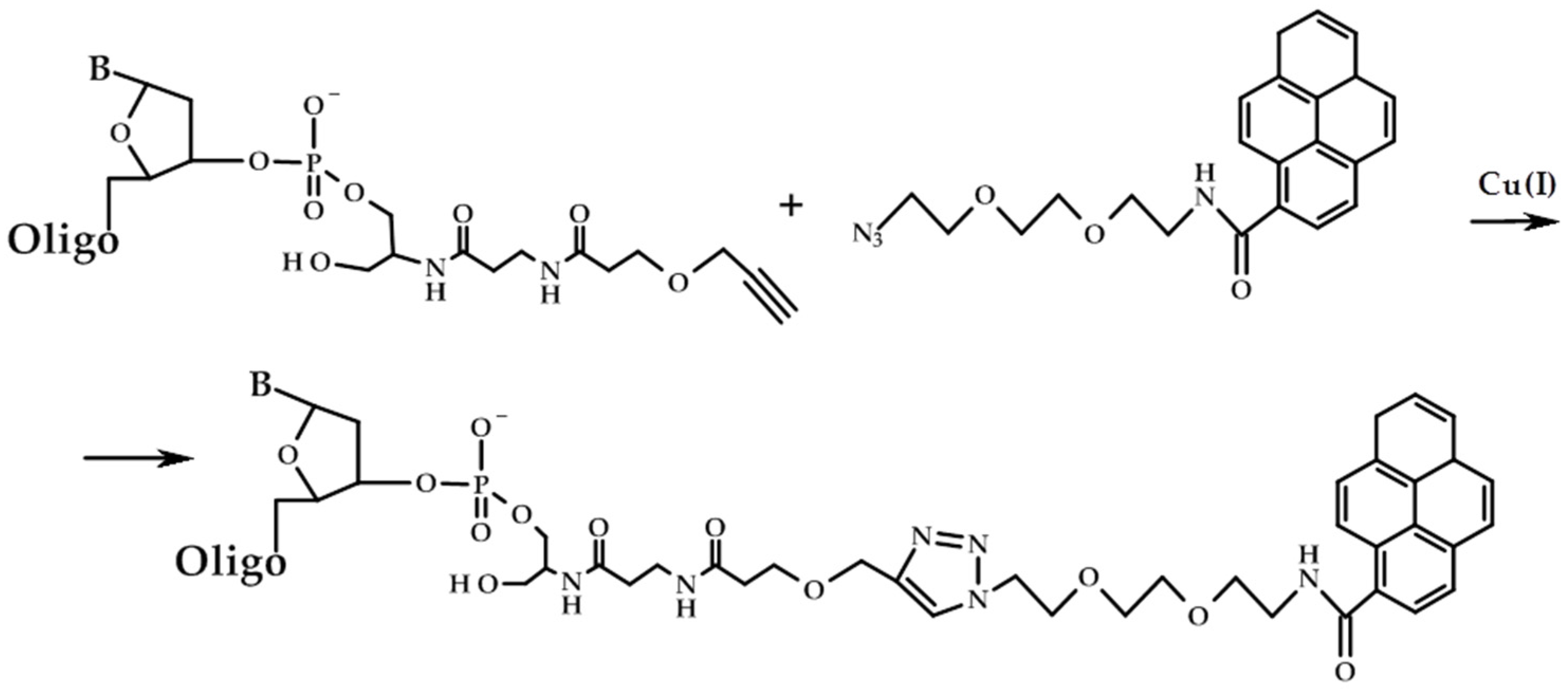
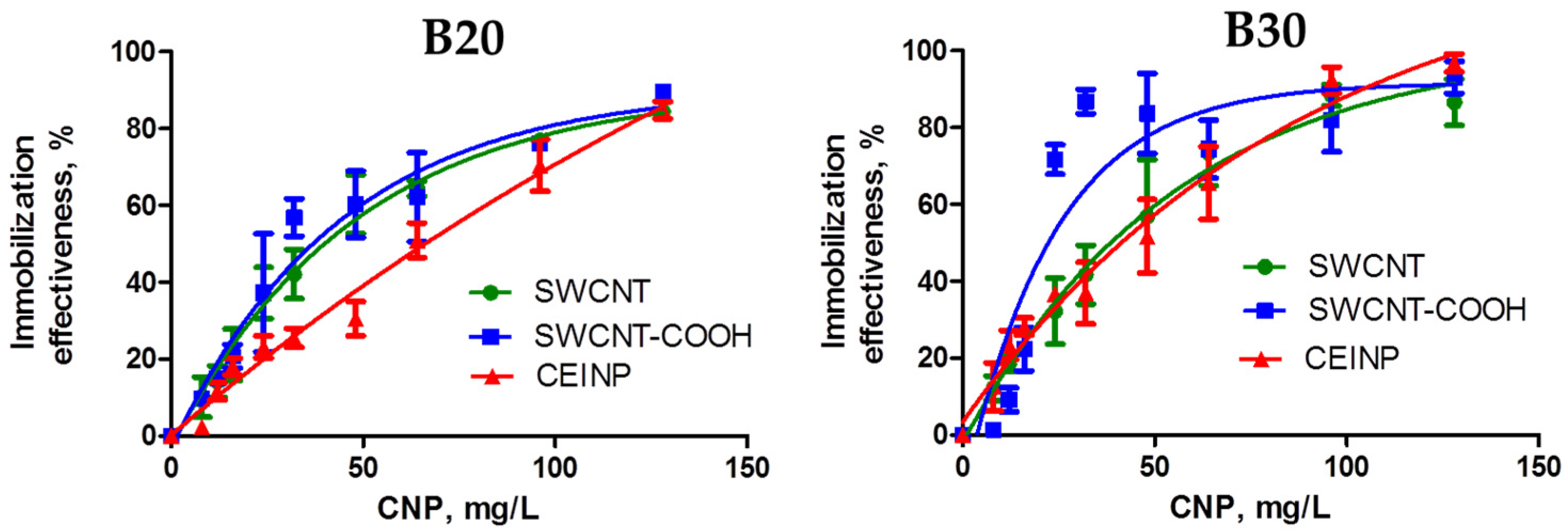
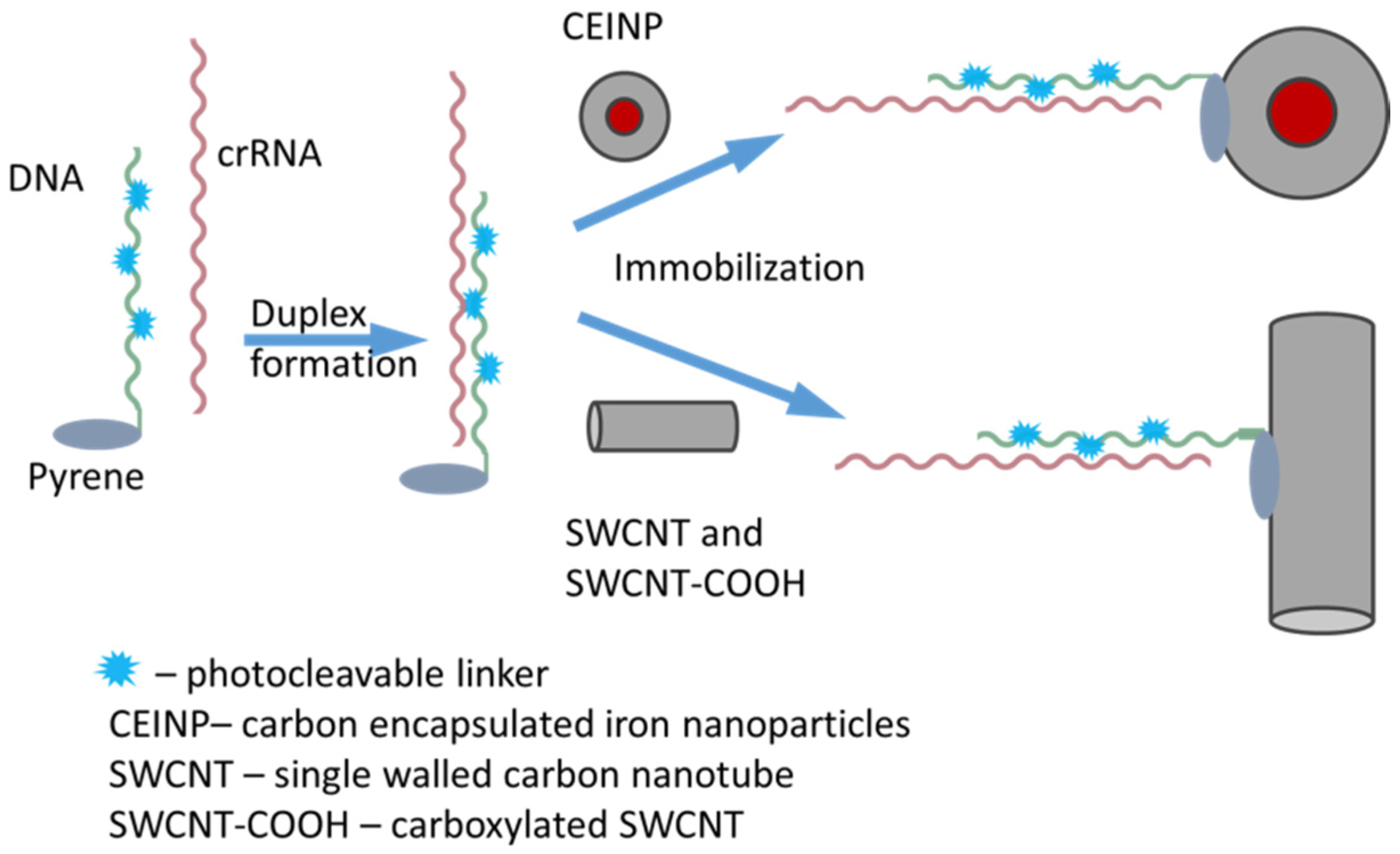
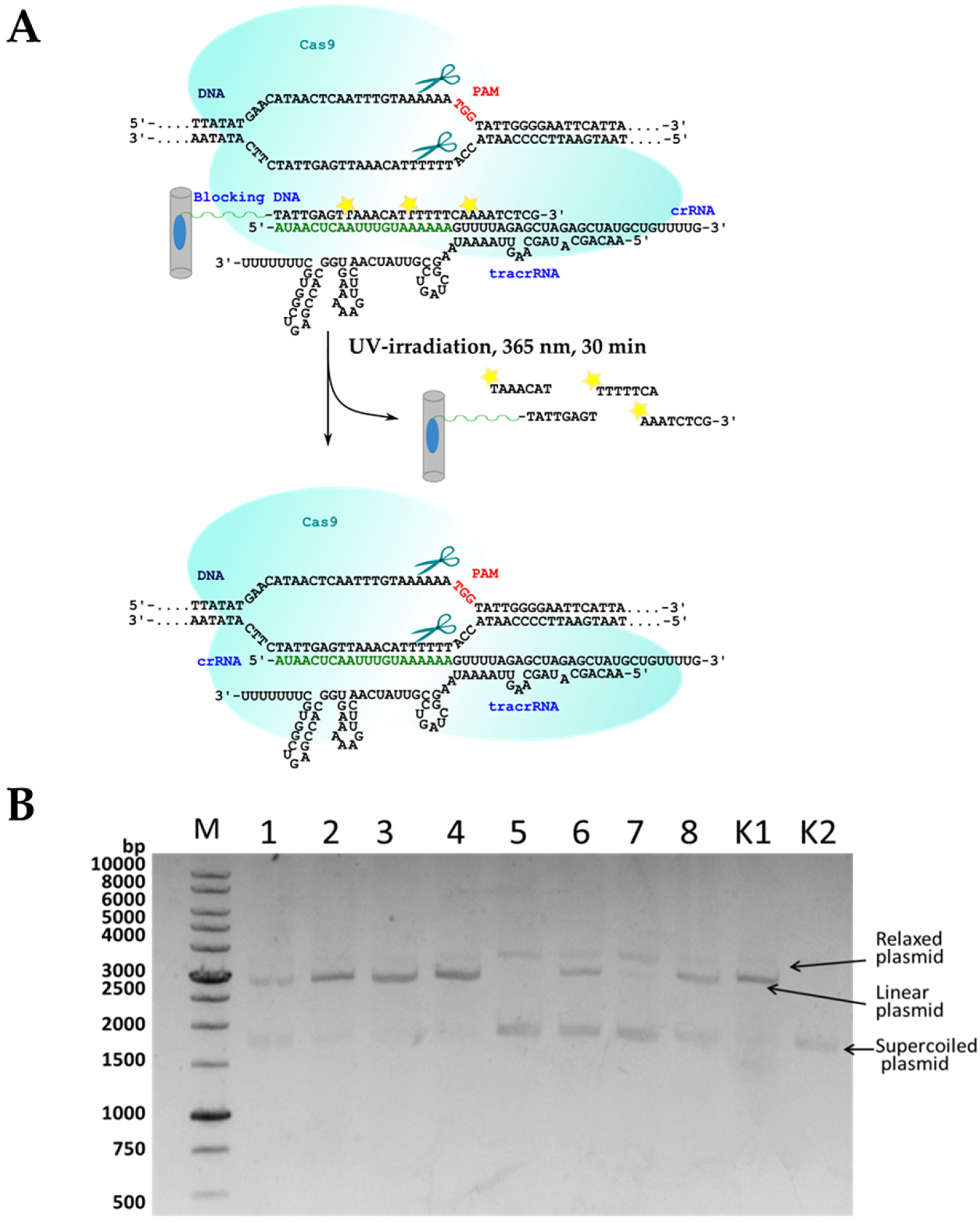
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Equipment
3.3. Physical Measurements
3.4. Synthesis of 2-O-4,4′-dimethoxytrityl-1-(2-nitrophenyl)-1,2-ethanediol (I)
3.5. Synthesis of 1-(2-cyano-N,N-diisopropyl-phosphoramidite)-2-O-4,4′-dimethoxytrityl-1-(2-nitrophenyl1,2-ethanediol (II)
3.6. Phosphoramidite Synthesis of Oligonucleotides
3.7. Synthesis of 3′-Pyrene Conjugates
3.8. Purification of Oligonucleotides and Their Conjugates
3.9. Oxidation of Carbon Nanotubes
3.10. Carbon Encapsulated Iron Nanoparticles Preparation
3.11. Adsorption Isotherms of Pyrene Conjugates of Oligonucleotides onto SWCNTs
3.12. Immobilization of crRNA on CNP Surface
3.13. The Release of Fluorescent crRNA_Flu Immobilized on CNP Surface
3.14. Cleavage of Plasmid DNA by the Cas9 Protein in the Presence of Immobilized Guide RNAs
3.15. The Efficiency of the Plasmid Cleavage
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Newsom, S.; Parameshwaran, H.P.; Martin, L.; Rajan, R. The CRISPR-Cas Mechanism for Adaptive Immunity and Alternate Bacterial Functions Fuels Diverse Biotechnologies. Front. Cell. Infect. Microbiol. 2021, 10, 898. [Google Scholar] [CrossRef]
- Hossain, M.A. CRISPR-Cas9: A fascinating journey from bacterial immune system to human gene editing. In Progress in Molecular Biology and Translational Science V. 178 Advances in CRISPR/Cas and Related Technologies; Ghosh, D., Ed.; Elsevier Inc.: Cambridge, MA, USA; San Diego, CA, USA; Oxford, UK; London, UK, 2021; pp. 63–83. [Google Scholar]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Brown, W.; Zhou, W.; Deiters, A. Regulating CRISPR/Cas9 Function through Conditional Guide RNA Control. ChemBioChem 2021, 22, 63–72. [Google Scholar] [CrossRef]
- Wu, H.; Wang, F.; Jiang, J. Inducible CRISPR-dCas9 Transcriptional Systems for Sensing and Genome Regulation. ChemBioChem 2021, 22, 1894–1900. [Google Scholar] [CrossRef]
- Zhou, X.X.; Zou, X.; Chung, H.K.; Gao, Y.; Liu, Y.; Qi, L.S.; Lin, M.Z. A Single-Chain Photoswitchable CRISPR-Cas9 Architecture for Light-Inducible Gene Editing and Transcription. ACS Chem. Biol. 2018, 13, 443–448. [Google Scholar] [CrossRef] [Green Version]
- Gjaltema, R.A.F.; Schulz, E.G. CRISPR/dCas9 Switch Systems for Temporal Transcriptional Control. In Epigenome Editing; Humana Press: New York, NY, USA, 2018; pp. 167–185. [Google Scholar]
- Renzl, C.; Kakoti, A.; Mayer, G. Aptamer-Mediated Reversible Transactivation of Gene Expression by Light. Angew. Chem. Int. Ed. 2020, 59, 22414–22418. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, R.S.; He, S.; Nihongaki, Y.; Li, X.; Razavi, S.; Wu, B.; Ha, T. Very fast CRISPR on demand. Science 2020, 368, 1265–1269. [Google Scholar] [CrossRef]
- Zhou, W.; Brown, W.; Bardhan, A.; Delaney, M.; Ilk, A.S.; Rauen, R.R.; Kahn, S.I.; Tsang, M.; Deiters, A. Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light-Activated Guide RNA. Angew. Chem. Int. Ed. 2020, 59, 8998–9003. [Google Scholar] [CrossRef]
- Zhang, Y.; Ling, X.; Su, X.; Zhang, S.; Wang, J.; Zhang, P.; Feng, W.; Zhu, Y.Y.; Liu, T.; Tang, X. Optical Control of a CRISPR/Cas9 System for Gene Editing by Using Photolabile crRNA. Angew. Chem. Int. Ed. 2020, 59, 20895–20899. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Xie, F.; Lin, J.; Xu, L. Photocontrol of CRISPR/Cas9 function by site-specific chemical modification of guide RNA. Chem. Sci. 2020, 11, 11478–11484. [Google Scholar] [CrossRef]
- Akhmetova, E.A.; Golyshev, V.M.; Vokhtantcev, I.P.; Meschaninova, M.I.; Venyaminova, A.G.; Novopashina, D.S. Photoactivatable CRISPR/Cas9 System. Russ. J. Bioorg. Chem. 2021, 47, 496–504. [Google Scholar] [CrossRef]
- Jain, P.K.; Ramanan, V.; Schepers, A.G.; Dalvie, N.S.; Panda, A.; Fleming, H.E.; Bhatia, S.N. Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angew. Chem. Int. Ed. 2016, 55, 12440–12444. [Google Scholar] [CrossRef]
- Alghuthaymi, M.A.; Ahmad, A.; Khan, Z.; Khan, S.H.; Ahmed, F.K.; Faiz, S.; Nepovimova, E.; Kuča, K.; Abd-Elsalam, K.A. Exosome/Liposome-like Nanoparticles: New Carriers for CRISPR Genome Editing in Plants. Int. J. Mol. Sci. 2021, 22, 7456. [Google Scholar] [CrossRef]
- Alimi, L.O.; Alyami, M.Z.; Chand, S.; Baslyman, W.; Khashab, N.M. Coordination-based self-assembled capsules (SACs) for protein, CRISPR–Cas9, DNA and RNA delivery. Chem. Sci. 2021, 12, 2329–2344. [Google Scholar] [CrossRef]
- Tao, Y.; Yi, K.; Hu, H.; Shao, D.; Li, M. Coassembly of nucleus-targeting gold nanoclusters with CRISPR/Cas9 for simultaneous bioimaging and therapeutic genome editing. J. Mater. Chem. B 2021, 9, 94–100. [Google Scholar] [CrossRef]
- Zhuang, J.; Tan, J.; Wu, C.; Zhang, J.; Liu, T.; Fan, C.; Li, J.; Zhang, Y. Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy. Nucleic Acids Res. 2020, 48, 8870–8882. [Google Scholar] [CrossRef]
- Gong, Y.; Tian, S.; Xuan, Y.; Zhang, S. Lipid and polymer mediated CRISPR/Cas9 gene editing. J. Mater. Chem. B 2020, 8, 4369–4386. [Google Scholar] [CrossRef]
- Kuhn, J.; Lin, Y.; Krhac Levacic, A.; Al Danaf, N.; Peng, L.; Höhn, M.; Lamb, D.C.; Wagner, E.; Lächelt, U. Delivery of Cas9/sgRNA Ribonucleoprotein Complexes via Hydroxystearyl Oligoamino Amides. Bioconjug. Chem. 2020, 31, 729–742. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef] [Green Version]
- Falato, L.; Gestin, M.; Langel, Ü. Cell-Penetrating Peptides Delivering siRNAs: An Overview. In Design and Delivery of SiRNA Therapeutics; Humana: New York, NY, USA, 2021; pp. 329–352. [Google Scholar]
- Taylor, R.E.; Zahid, M. Cell Penetrating Peptides, Novel Vectors for Gene Therapy. Pharmaceutics 2020, 12, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Vanegas, J.D.; Cruz, J.C.; Reyes, L.H. Delivery Systems for Nucleic Acids and Proteins: Barriers, Cell Capture Pathways and Nanocarriers. Pharmaceutics 2021, 13, 428. [Google Scholar] [CrossRef]
- Zare, H.; Ahmadi, S.; Ghasemi, A.; Ghanbari, M.; Rabiee, N.; Bagherzadeh, M.; Karimi, M.; Webster, T.J.; Hamblin, M.R.; Mostafavi, E. Carbon Nanotubes: Smart Drug/Gene Delivery Carriers. Int. J. Nanomed. 2021, 16, 1681–1706. [Google Scholar] [CrossRef] [PubMed]
- Debnath, S.K.; Srivastava, R. Drug Delivery With Carbon-Based Nanomaterials as Versatile Nanocarriers: Progress and Prospects. Front. Nanotechnol. 2021, 3, 15. [Google Scholar] [CrossRef]
- Apartsin, E.K.; Buyanova, M.Y.; Novopashina, D.S.; Ryabchikova, E.I.; Filatov, A.V.; Zenkova, M.A.; Venyaminova, A.G. Novel Multifunctional Hybrids of Single-Walled Carbon Nanotubes with Nucleic Acids: Synthesis and Interactions with Living Cells. ACS Appl. Mater. Interfaces 2014, 6, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Chen, B. A Review on Biomedical Applications of Single-Walled Carbon Nanotubes. Curr. Med. Chem. 2010, 17, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Hepel, M. Magnetic Nanoparticles for Nanomedicine. Magnetochemistry 2020, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Price, P.M.; Mahmoud, W.E.; Al-Ghamdi, A.A.; Bronstein, L.M. Magnetic Drug Delivery: Where the Field Is Going. Front. Chem. 2018, 6, 619. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.F.; Jang, B.; Issadore, D.; Tsourkas, A. Use of magnetic fields and nanoparticles to trigger drug release and improve tumor targeting. WIREs Nanomed. Nanobiotechnol. 2019, 11, e1571. [Google Scholar] [CrossRef]
- Mirvakili, S.M.; Langer, R. Wireless on-demand drug delivery. Nat. Electron. 2021, 4, 464–477. [Google Scholar] [CrossRef]
- Wu, Y.; Phillips, J.A.; Liu, H.; Yang, R.; Tan, W. Carbon Nanotubes Protect DNA Strands during Cellular Delivery. ACS Nano 2008, 2, 2023–2028. [Google Scholar] [CrossRef] [Green Version]
- Apartsin, E.K.; Buyanova, M.Y.; Novopashina, D.S.; Venyaminova, A.G. Hybrids of siRNA with Carbon Nanotubes as RNA Interference Instruments. In Nanobiophysics-Fundamentals and Applications; Karachevtsev, V., Ed.; Pan Stanford Publishing: Singapore, 2016; pp. 33–58. [Google Scholar]
- Neagoe, I.B.; Braicu, C.; Matea, C.; Bele, C.; Florin, G.; Gabriel, K.; Veronica, C.; Irimie, A. Efficient siRNA Delivery System Using Carboxilated Single-Wall Carbon Nanotubes in Cancer Treatment. J. Biomed. Nanotechnol. 2012, 8, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Charbgoo, F.; Behmanesh, M.; Nikkhah, M.; Kane, E.G. RNAi mediated gene silencing of ITPA using a targeted nanocarrier: Apoptosis induction in SKBR3 cancer cells. Clin. Exp. Pharmacol. Physiol. 2017, 44, 888–894. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Lin, I.-J.; Chen, C.-C.; Hsu, Y.-C.; Chang, C.-C.; Lee, M.-J. Delivery of small interfering RNAs in human cervical cancer cells by polyethylenimine-functionalized carbon nanotubes. Nanoscale Res. Lett. 2013, 8, 267. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Hu, Y.; Zhao, J.-J. Repression of Multiple Myeloma Cell Growth In Vivo by Single-wall Carbon Nanotube (SWCNT)-delivered MALAT1 Antisense Oligos. J. Vis. Exp. 2018, 142, e58598. [Google Scholar] [CrossRef] [PubMed]
- Pinals, R.L.; Yang, D.; Rosenberg, D.J.; Chaudhary, T.; Crothers, A.R.; Iavarone, A.T.; Hammel, M.; Landry, M.P. Quantitative Protein Corona Composition and Dynamics on Carbon Nanotubes in Biological Environments. Angew. Chem. Int. Ed. 2020, 59, 23668–23677. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, C.; Lin, Q.; Li, N.; Huang, Z.-B.; Zhao, M.; Fu, X.-Z.; Wang, G.-X.; Zhu, B. Single-walled carbon nanotubes as delivery vehicles enhance the immunoprotective effect of an immersion DNA vaccine against infectious spleen and kidney necrosis virus in mandarin fish. Fish Shellfish Immunol. 2020, 97, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Apartsin, E.K.; Novopashina, D.S.; Nastaushev, Y.V.; Ven’yaminova, A.G. Fluorescently labeled single-walled carbon nanotubes and their hybrids with oligonucleotides. Nanotechnol. Russ. 2012, 7, 99–109. [Google Scholar] [CrossRef]
- Permyakova, E.S.; Novopashina, D.S.; Venyaminova, A.G.; Apartsin, E.K. Non-covalent anchoring of oligonucleotides on single-walled carbon nanotubes via short bioreducible linker. RSC Adv. 2017, 7, 29212–29217. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, L.; Xue, X.; Ge, G.; Liang, X. Enhanced dispersibility and cellular transmembrane capability of single-wall carbon nanotubes by polycyclic organic compounds as chaperon. Nanoscale 2012, 4, 3983. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, S.; Zhou, W.; Xu, J.; Li, Y. Spectroscopic Evidence and Molecular Simulation Investigation of the π–π Interaction Between Pyrene Molecules and Carbon Nanotubes. J. Nanosci. Nanotechnol. 2007, 7, 2366–2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehli, C.; Rahman, G.M.A.; Jux, N.; Balbinot, D.; Guldi, D.M.; Paolucci, F.; Marcaccio, M.; Paolucci, D.; Melle-Franco, M.; Zerbetto, F.; et al. Interactions in Single Wall Carbon Nanotubes/Pyrene/Porphyrin Nanohybrids. J. Am. Chem. Soc. 2006, 128, 11222–11231. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, T.; Li, J.; Guo, Z.-X.; Dai, L.; Zhang, D.; Zhu, D. Self-assembly of gold nanoparticles to carbon nanotubes using a thiol-terminated pyrene as interlinker. Chem. Phys. Lett. 2003, 367, 747–752. [Google Scholar] [CrossRef]
- Anders, C.; Jinek, M. In Vitro Enzymology of Cas9. Methods Enzymol. 2014, 546, 1–20. [Google Scholar] [PubMed] [Green Version]
- Ji, Y.; Yang, J.; Wu, L.; Yu, L.; Tang, X. Photochemical Regulation of Gene Expression Using Caged siRNAs with Single Terminal Vitamin E Modification. Angew. Chem. Int. Ed. 2016, 55, 2152–2156. [Google Scholar] [CrossRef]
- Tang, X.; Su, M.; Yu, L.; Lv, C.; Wang, J.; Li, Z. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides. Nucleic Acids Res. 2010, 38, 3848–3855. [Google Scholar] [CrossRef] [Green Version]
- Gromov, A.; Dittmer, S.; Svensson, J.; Nerushev, O.A.; Perez-García, S.A.; Licea-Jiménez, L.; Rychwalski, R.; Campbell, E.E.B. Covalent amino-functionalisation of single-wall carbon nanotubes. J. Mater. Chem. 2005, 15, 3334. [Google Scholar] [CrossRef]
- Smovzh, D.V.; Sakhapov, S.Z.; Zaikovskii, A.V.; Novopashin, S.A. Morphology of aluminium oxide nanostructures after calcination of arc discharge Al–C soot. Ceram. Int. 2015, 41, 8814–8819. [Google Scholar] [CrossRef]
- Novopashin, S.A.; Serebryakova, M.A.; Zaikovskii, A.V. Morphology, Chemical Composition, and Magnetic Properties of Arc Discharge Fe–C Soot. In Proceedings of the 3rd International Multidisciplinary Microscopy and Microanalysis Congress (InterM), Oludeniz, Turkey, 19–23 October 2015; Springer Proceedings in Physics. Oral, A., Bahsi Oral, Z., Eds.; Springer: Cham, Switzerland, 2017; pp. 149–155. [Google Scholar]
- Shubsda, M.F.; Goodisman, J.; Dabrowiak, J.C. Quantitation of ethidium-stained closed circular DNA in agarose gels. J. Biochem. Biophys. Methods 1997, 34, 73–79. [Google Scholar] [CrossRef] [Green Version]


| Name | Sequence, 5′-3′ |
|---|---|
| B20 | 5′-TTTTTTACAAATTGAGTTATCC-Pyr |
| B20_PL | 5′-TTTTTT-PL-ACAAA-PL-TTGAG-PL-TTATCC-Pyr |
| B30 | 5′-GCTCTAAAACTTTTTTACAAATTGAGTTAT-Pyr |
| B30_PL | 5′-GCTCTAAA-PL-ACTTTTT-PL-TACAAAT-PL-TGAGTTAT-Pyr |
| crRNA_Flu | 5′-AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAUGCUGUUUUG-Flu |
| crRNA | 5′-AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAUGCUGUUUUG |
| tracrRNA | 5′-AACAGCAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUUUU |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semikolenova, O.; Sakovina, L.; Akhmetova, E.; Kim, D.; Vokhtantsev, I.; Golyshev, V.; Vorobyeva, M.; Novopashin, S.; Novopashina, D. Photoactivatable nanoCRISPR/Cas9 System Based on crRNA Reversibly Immobilized on Carbon Nanoparticles. Int. J. Mol. Sci. 2021, 22, 10919. https://doi.org/10.3390/ijms222010919
Semikolenova O, Sakovina L, Akhmetova E, Kim D, Vokhtantsev I, Golyshev V, Vorobyeva M, Novopashin S, Novopashina D. Photoactivatable nanoCRISPR/Cas9 System Based on crRNA Reversibly Immobilized on Carbon Nanoparticles. International Journal of Molecular Sciences. 2021; 22(20):10919. https://doi.org/10.3390/ijms222010919
Chicago/Turabian StyleSemikolenova, Olga, Lubov Sakovina, Elizaveta Akhmetova, Daria Kim, Ivan Vokhtantsev, Victor Golyshev, Mariya Vorobyeva, Sergey Novopashin, and Darya Novopashina. 2021. "Photoactivatable nanoCRISPR/Cas9 System Based on crRNA Reversibly Immobilized on Carbon Nanoparticles" International Journal of Molecular Sciences 22, no. 20: 10919. https://doi.org/10.3390/ijms222010919







