ATG 4B Serves a Crucial Role in RCE-4-Induced Inhibition of the Bcl-2–Beclin 1 Complex in Cervical Cancer Ca Ski Cells
Abstract
:1. Introduction
2. Results
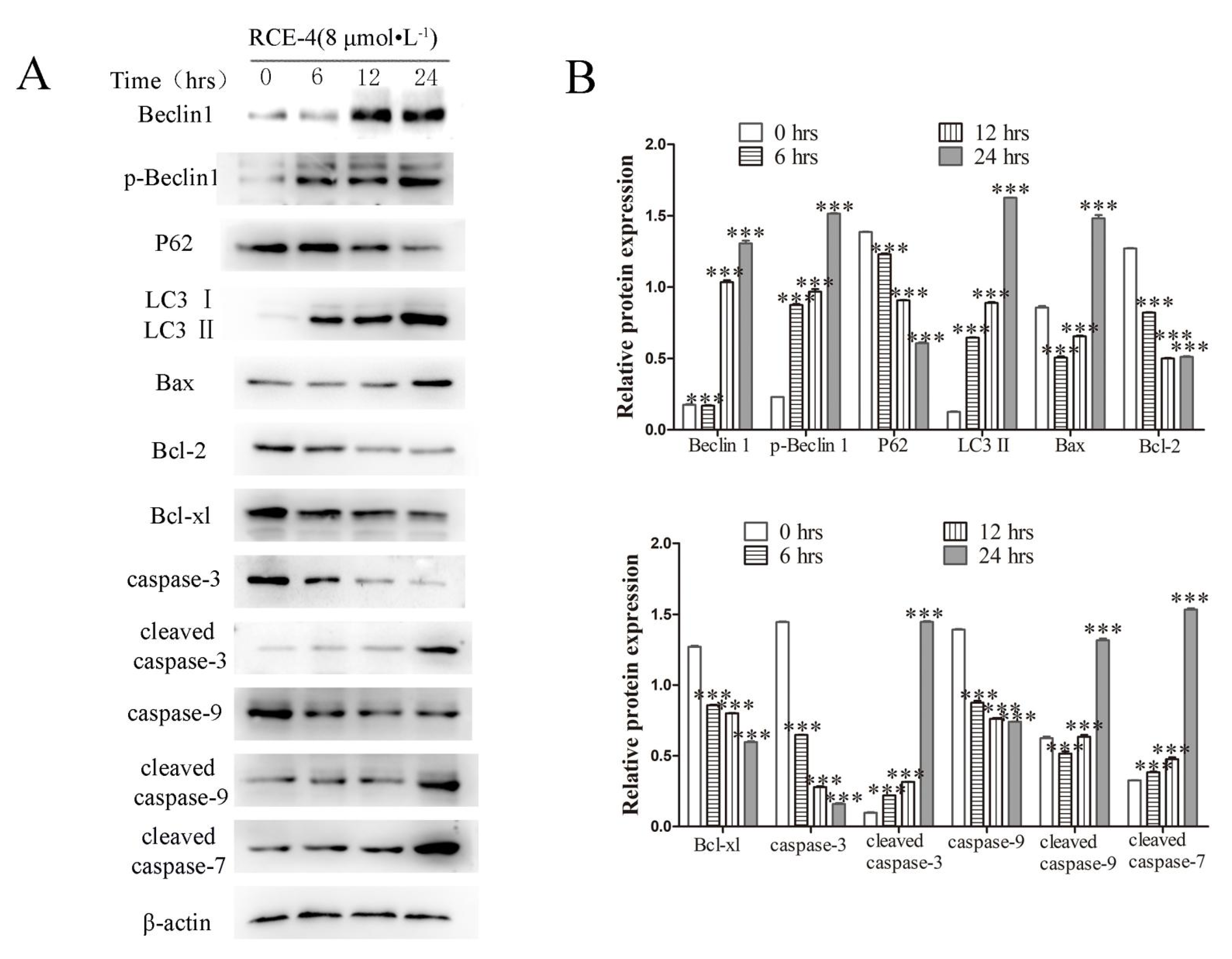
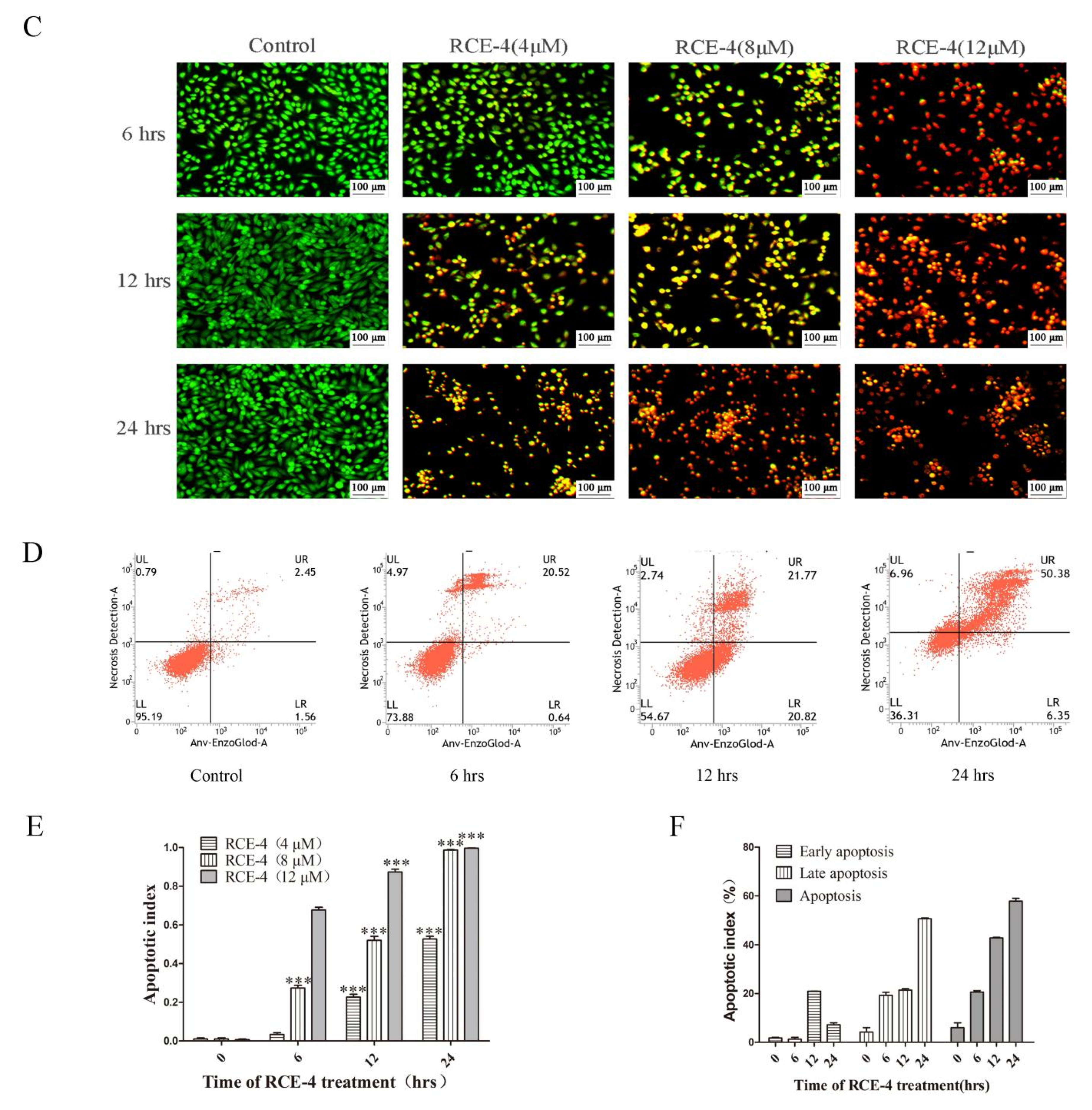
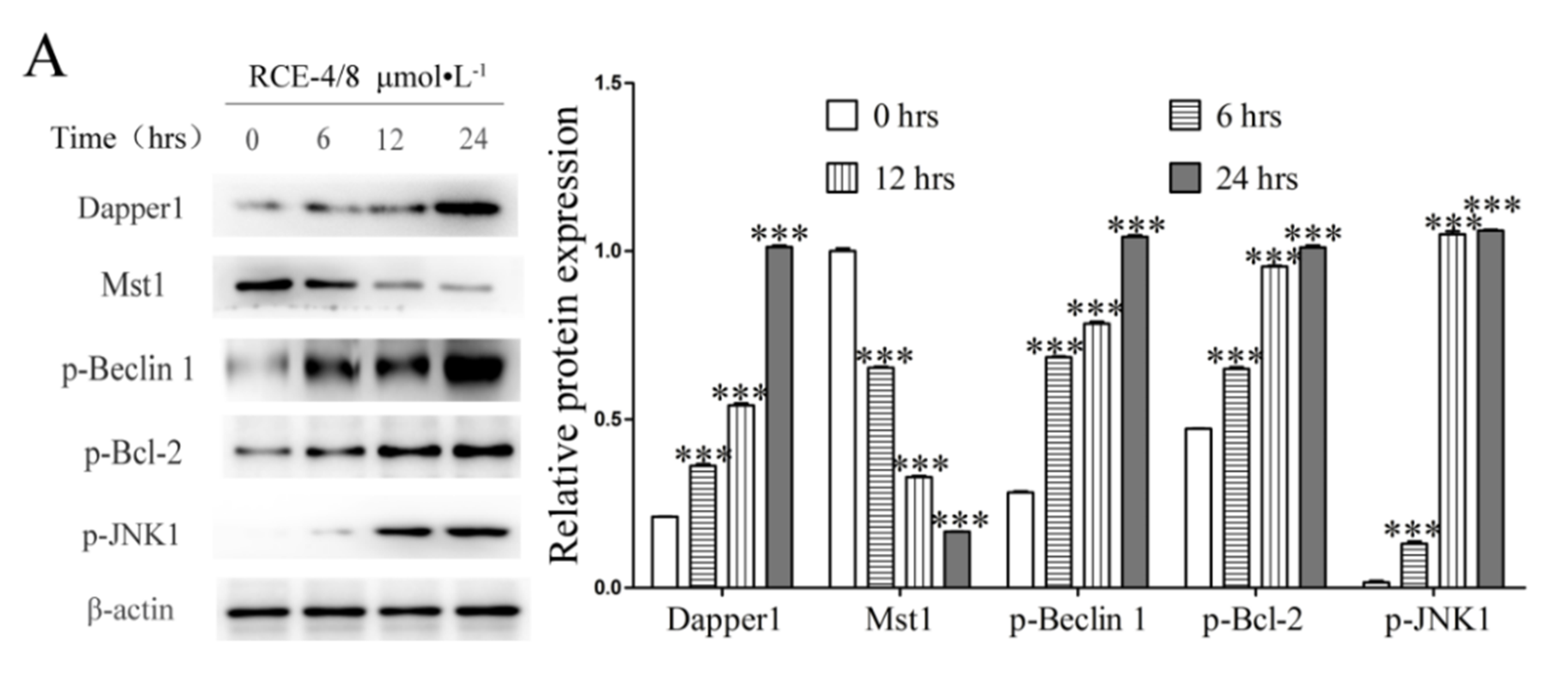
2.1. RCE-4 Induces Time- and Concentration-Dependent Apoptosis and Autophagy in Ca Ski Cells
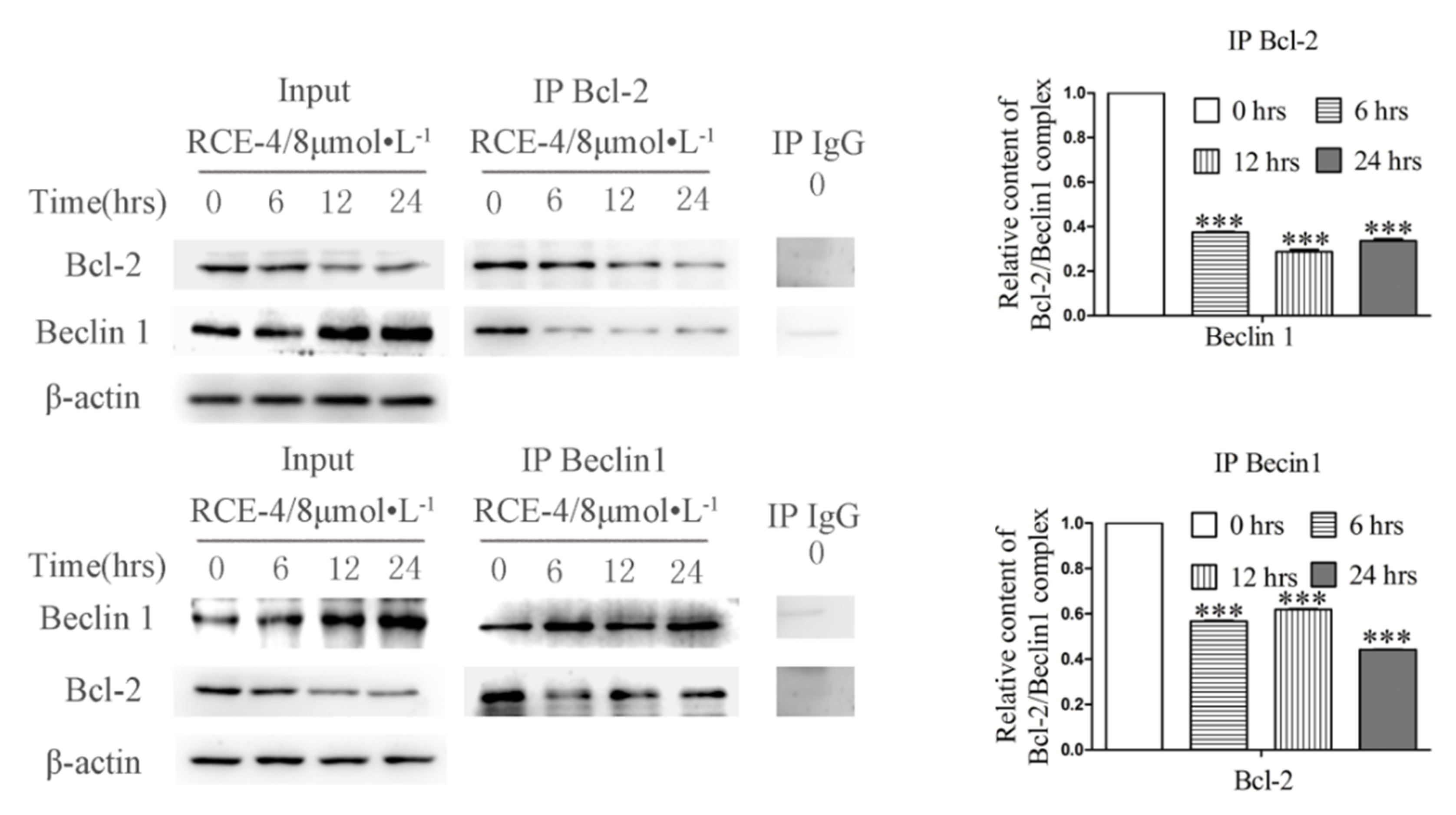
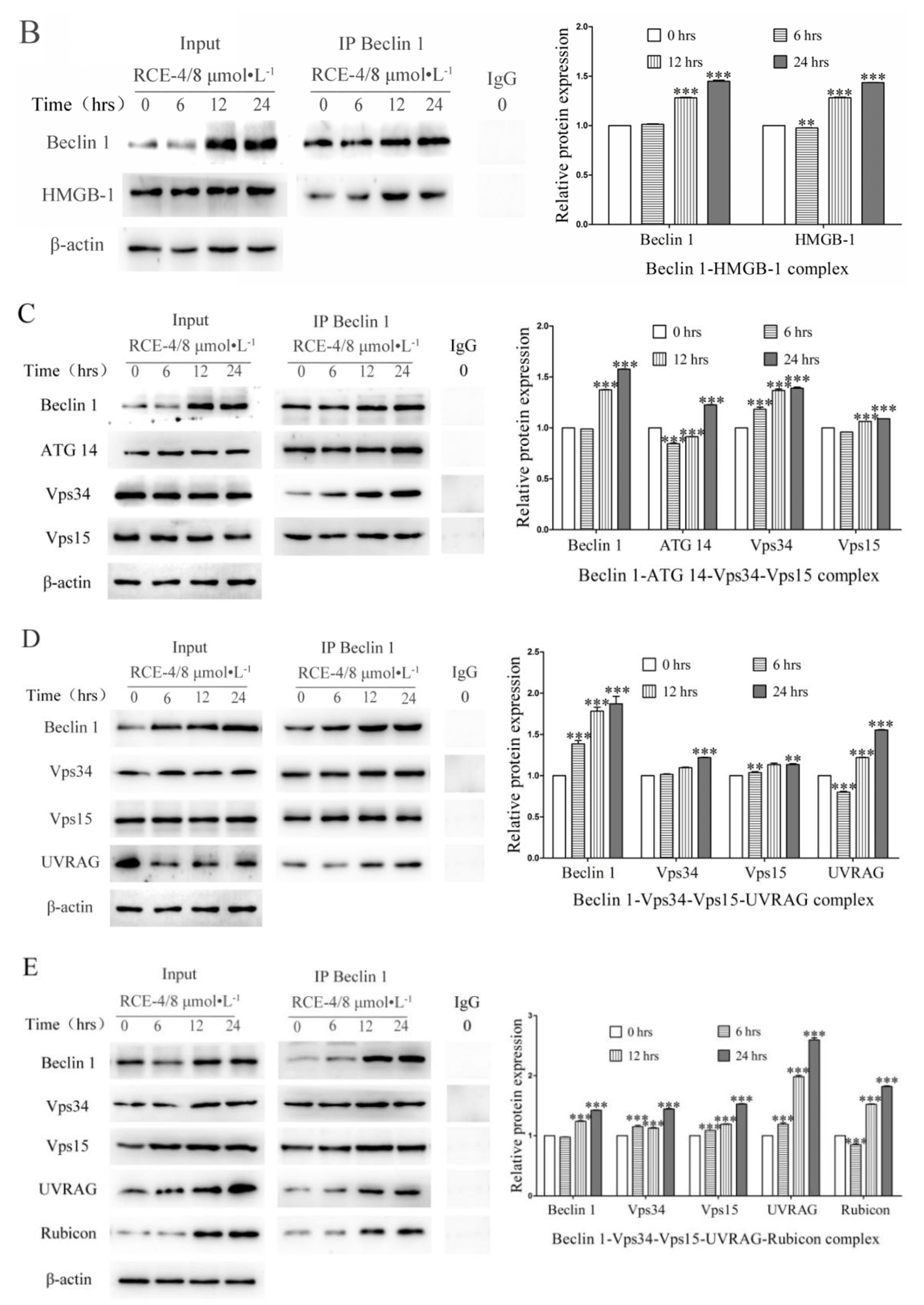
2.2. RCE-4 Inhibits the Formation of Bcl-2–Beclin 1 Complex in Ca Ski Cells
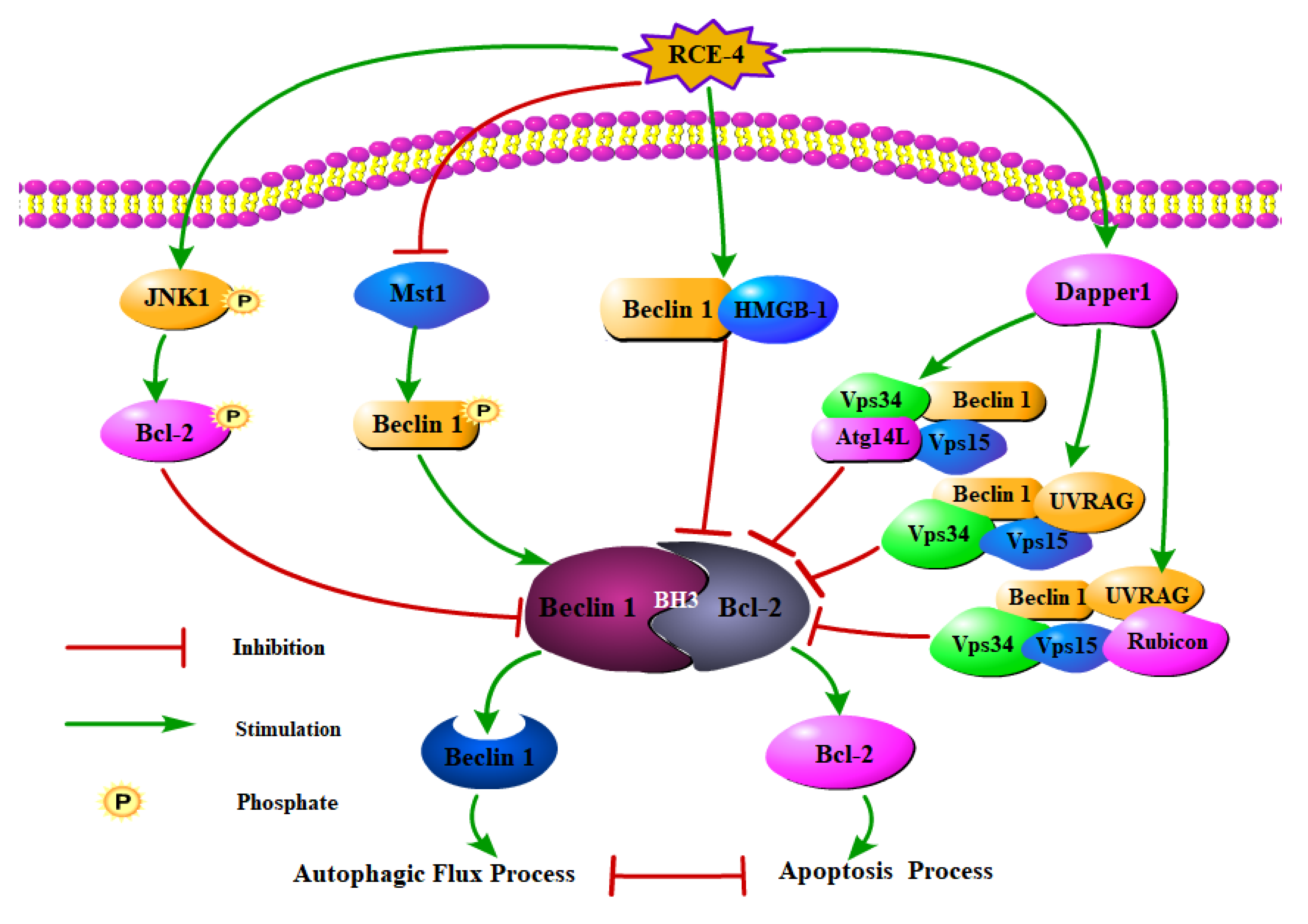
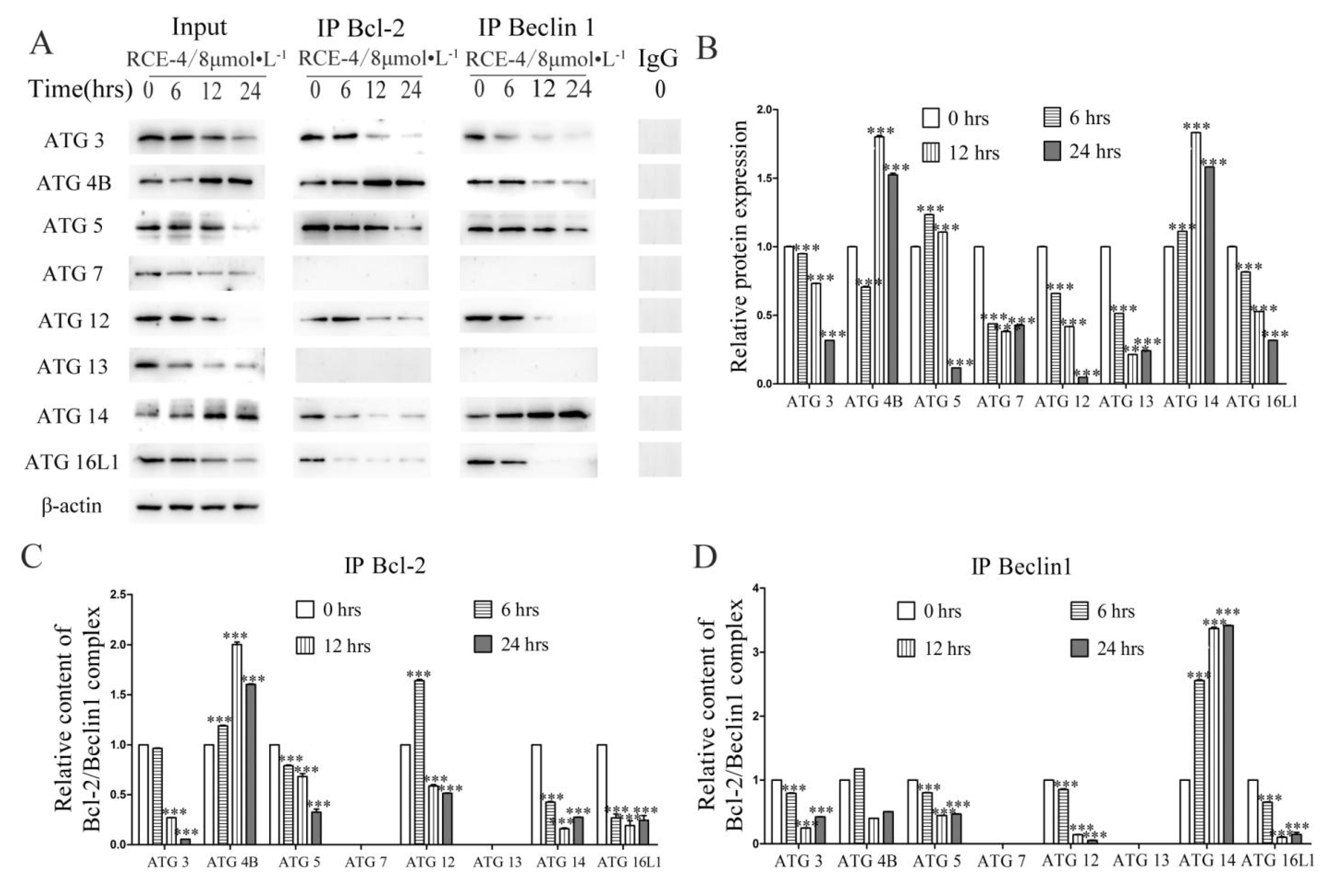
2.3. RCE-4 Inhibits the Formation of Bcl-2–Beclin 1 Complex via Various Pathways
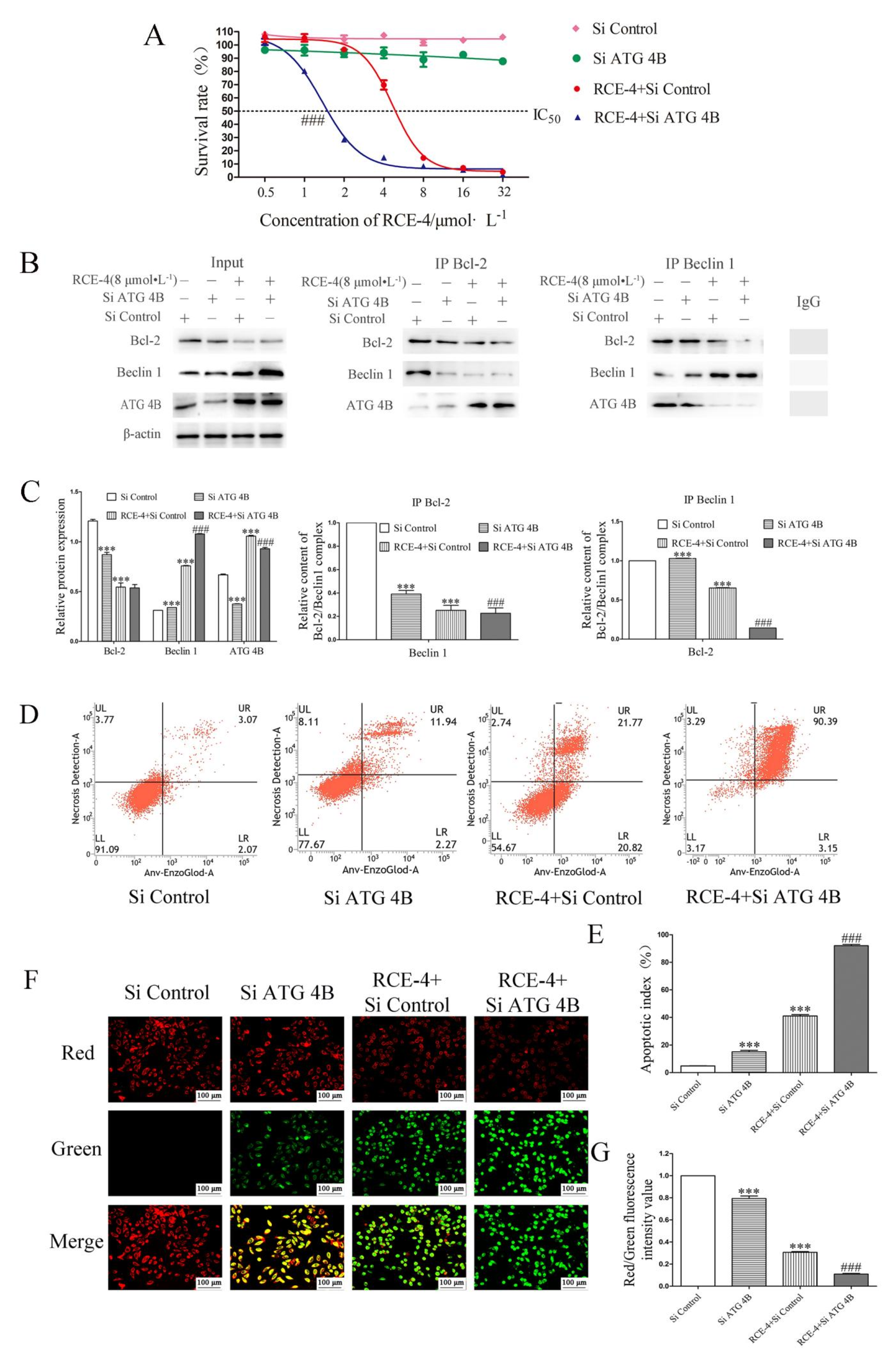
2.4. ATG 4B Plays a Critical Role in the Inhibition of Bcl-2–Beclin 1 Complex Induced by RCE-4
2.5. RCE-4 Combined with ATG siRNA Enhances the Sensitivity to Ca Ski Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reagent and Antibodies
4.3. Cytotoxicity Assay
4.4. Flow Cytometry Assay
4.5. Acridine Orange/Ethidium Bromide (AO/EB) Double Fluorescent Staining
4.6. MMP Assay
4.7. Western Blotting
4.8. Plasmid Small Interfering (si)RNA Transfection
4.9. Co-Immunoprecipitation (Co-IP)
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, Z.; Ma, D. The precision prevention and therapy of HPV-related cervical cancer: New concepts and clinical implications. Cancer Med. 2018, 7, 5217–5236. [Google Scholar] [CrossRef]
- Xiang, W.; Zhang, R.; Jin, G.; Tian, L.; Cheng, F.; Wang, J.; Xing, X.; Xi, W.; Tang, S.; Chen, J. RCE-4, a potential anti-cervical cancer drug isolated from Reineckia carnea, induces autophagy via the dual blockade of PI3K and ERK pathways in cervical cancer CaSki cells. Int. J. Mol. Med. 2019, 45, 245–254. [Google Scholar] [CrossRef]
- Bai, C.; Yang, X.; Zou, K.; He, H.; Wang, J.; Qin, H.; Yu, X.; Liu, C.; Zheng, J.; Cheng, F.; et al. Anti-proliferative effect of RCE-4 from Reineckia carnea on human cervical cancer HeLa cells by inhibiting the PI3K/Akt/mTOR signaling pathway and NF-κB activation. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2016, 389, 573–584. [Google Scholar] [CrossRef]
- Wang, G.; Huang, W.; He, H.; Fu, X.; Wang, J.; Zou, K.; Chen, J. Growth inhibition and apoptosis-inducing effect on human cancer cells by RCE-4, a spirostanol saponin derivative from natural medicines. Int. J. Mol. Med. 2012, 31, 219–224. [Google Scholar] [CrossRef]
- Yan, W.; Zou, K.; He, H.; Zhang, Y.; Li, X.; LI, X.; Yang, X.; Wang, J.; Deng, Z. Effects of steroidal saponin RCE-4 from Reineckia carnea (Andr.) Kunth on Ras/Erk and p16/cyclin D1/CDK4 signaling pathways in the human cervix cancer Ca Ski cells. Chin. J. Clin. Pharmacol. Ther. 2018, 23, 247–254. [Google Scholar]
- Yang, X.J.; Bai, C.H.; Zou, K.; He, H.B.; Yu, X.Q. Steroidal saponin RCE-4 from Reineckia carnea (Andr.) Kunth inhibits growth of human cervical cancer xenograft in nude mice. J. Third Mil. Med. Univ. 2016, 5, 476–482. (In Chinese) [Google Scholar]
- Hubei Institute for Drug Control. Quality Standards for Traditional Chinese Medicines in Hubei Province; Hubei Science and Technology Press: Wuhan, China, 2009. (In Chinese) [Google Scholar]
- Ke, B.; Tian, M.; Li, J.; Liu, B.; He, G. Targeting programmed cell death using small-molecule compounds to improve potential cancer therapy. Med. Res. Rev. 2016, 36, 983–1035. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in Human Health and Disease. N. Engl. J. Med. 2013, 368, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, R.M. Apoptosis and Caspases in Neurodegenerative Diseases. N. Engl. J. Med. 2003, 348, 1365–1375. [Google Scholar] [CrossRef]
- Mishra, A.P.; Salehi, B.; Sharifi-Rad, M.; Pezzani, R.; Kobarfard, F.; Sharifi-Rad, J.; Nigam, M. Programmed Cell Death, from a Cancer Perspective: An Overview. Mol. Diagn. Ther. 2018, 22, 281–295. [Google Scholar] [CrossRef]
- Gordy, C.; He, Y. The crosstalk between autophagy and apoptosis: Where does this lead? Protein Cell 2012, 3, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Jiang, M.; Chen, W.; Zhao, T.; Wei, Y. Faculty Opinions recommendation of Cancer and ER stress: Mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed. Pharmacother. 2020, 118, 109249. [Google Scholar] [CrossRef]
- Kapuy, O.; Vinod, P.K.; Mandl, J.; Bánhegyi, G. A cellular stress-directed bistable switch controls the crosstalk between autophagy and apoptosis. Mol. BioSyst. 2013, 9, 296–306. [Google Scholar] [CrossRef] [Green Version]
- Decuypere, J.-P.; Parys, J.B.; Bultynck, G. Regulation of the Autophagic Bcl-2/Beclin 1 Interaction. Cells 2012, 1, 284–312. [Google Scholar] [CrossRef]
- Marquez, R.T.; Xu, L. Bcl-2-Beclin 1 complex: Multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am. J. Cancer Res. 2012, 2, 214–221. [Google Scholar] [PubMed]
- Nopparat, C.; Porter, J.E.; Ebadi, M.; Govitrapong, P. 1-Methyl-4-phenylpyridinium-induced cell death via autophagy through a Bcl-2/Beclin 1 complex-dependent pathway. Neurochem. Res. 2014, 39, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Bishayee, K.; Habib, K.; Sadra, A.; Huh, S.-O. 18α-Glycyrrhetinic acid lethality for neuroblastoma cells via de-regulating the Beclin-1/Bcl-2 complex and inducing apoptosis. Biochem. Pharmacol. 2016, 117, 97–112. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhu, H.; Li, H.; Zou, M.-H.; Xie, Z. Dissociation of Bcl-2-Beclin1 complex by activated AMPK enhances cardiac autophagy and protects against cardiomyocyte apoptosis in diabetes. Diabetes 2012, 62, 1270–1281. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Wang, J.; Hou, J.; Fu, J.; Chang, D.; Bensoussan, A.; Liu, J. Ginsenoside Rg1 protects starving H9c2 cells by dissociation of Bcl-2-Beclin1 complex. BMC Complement. Altern. Med. 2016, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ke, D.; Ji, L.; Wang, Y.; Fu, X.; Chen, J.; Wang, F.; Zhao, D.; Xue, Y.; Lan, X.; Hou, J. JNK1 regulates RANKL-induced osteoclastogenesis via activation of a novel Bcl-2-Beclin1-autophagy pathway. FASEB J. 2019, 33, 11082–11095. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-Mediated Phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maejima, Y.; Kyoi, S.; Zhai, P.; Liu, T.; Li, H.; Ivessa, A.; Sciarretta, S.; Del Re, D.P.; Zablocki, D.K.; Hsu, C.-P.; et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 2013, 19, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Cao, W.; Li, W.; Gao, C.; Qi, Z.; Zhao, Y.; Du, J.; Xue, H.; Peng, J.; Wen, J.; et al. Dapper1 promotes autophagy by enhancing the Beclin1-Vps34-Atg14L complex formation. Cell Res. 2014, 24, 912–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menzies, F.M.; Fleming, A.; Rubinsztein, D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015, 16, 345–357. [Google Scholar] [CrossRef]
- Li, Z.; Li, Q.; Lv, W.; Jiang, L.; Geng, C.; Yao, X.; Shi, X.; Liu, Y.; Cao, J. The interaction of Atg4B and Bcl-2 plays an important role in Cd-induced crosstalk between apoptosis and autophagy through disassociation of Bcl-2-Beclin1 in A549 cells. Free Radic. Biol. Med. 2019, 130, 576–591. [Google Scholar] [CrossRef]
- Wei, Y.; Sinha, S.C.; Levine, B. Dual Role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008, 4, 949–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Kang, R.; Livesey, K.M.; Cheh, C.-W.; Farkas, A.M.; Loughran, P.; Hoppe, G.; Bianchi, M.E.; Tracey, K.J.; Zeh, H.J.; et al. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 2010, 190, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Cheh, C.-W.; Livesey, K.M.; Liang, X.; Schapiro, N.E.; Benschop, R.; Sparvero, L.J.; Amoscato, A.; Tracey, K.J.; et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene 2010, 29, 5299–5310. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Xu, L.-H.; Xu, W.; Fang, J.-P.; Xu, H.-G. HMGB1 translocation is involved in the transformation of autophagy complexes and promotes chemoresistance in leukaemia. Int. J. Oncol. 2015, 47, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Obara, K.; Ohsumi, Y. Atg14: A Key Player in Orchestrating Autophagy. Int. J. Cell Biol. 2011, 2011, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Fernández, F.; Sebti, S.; Wei, Y.; Zou, Z.; Shi, M.; McMillan, K.L.; He, C.; Ting, T.; Liu, Y.; Chiang, W.-C.; et al. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018, 558, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, W.; Guo, X.; Ren, J.; Gao, A. The crosstalk between autophagy and apoptosis was mediated by phosphorylation of Bcl-2 and beclin1 in benzene-induced hematotoxicity. Cell Death Dis. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.; Karnak, D.; Xu, L. The Bcl-2-Beclin 1 interaction in (−)-gossypol-induced autophagy versus apoptosis in prostate cancer cells. Autophagy 2010, 6, 1201–1203. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of ATG proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Su, J.; Qian, H.; Guo, T. SLC27A4 regulate ATG4B activity and control reactions to chemotherapeutics-induced autophagy in human lung cancer cells. Tumor Biol. 2015, 37, 6943–6952. [Google Scholar] [CrossRef]
- Vezenkov, L.; Honson, N.S.; Kumar, N.S.; Bosc, D.; Kovacic, S.; Nguyen, T.G.; Pfeifer, T.A.; Young, R.N. Development of fluorescent peptide substrates and assays for the key autophagy-initiating cysteine protease enzyme, ATG4B. Bioorg. Med. Chem. 2015, 23, 3237–3247. [Google Scholar] [CrossRef]
- Zhang, L.; Li, J.; Ouyang, L.; Liu, B.; Cheng, Y. Unraveling the roles of Atg4 proteases from autophagy modulation to targeted cancer therapy. Cancer Lett. 2016, 373, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Betin, V.M.; MacVicar, T.D.; Parsons, S.F.; Anstee, D.J.; Lane, J.D. A cryptic mitochondrial targeting motif in Atg4D links caspase cleavage with mitochondrial import and oxidative stress. Autophagy 2012, 8, 664–676. [Google Scholar] [CrossRef] [Green Version]
- Betin, V.M.; Lane, J.D. Atg4D at the interface between autophagy and apoptosis. Autophagy 2009, 5, 1057–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyo, J.O.; Jang, M.H.; Kwon, Y.K.; Lee, H.J.; Jun, J.I.; Woo, H.N.; Cho, D.H.; Choi, B.; Lee, H.; Kim, J.H.; et al. Essential roles of Atg5 and FADD in autophagic cell death: Dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 2005, 280, 20722–20729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codogno, P.; Meijer, A.J. Atg5: More than an autophagy factor. Nat. Cell Biol. 2006, 8, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.; Zhao, Z.; Stephenson, L.M.; Cadwell, K.; Pua, H.H.; Lee, H.K.; Mizushima, N.; Iwasaki, A.; He, Y.-W.; Swat, W.; et al. The autophagy geneATG5plays an essential role in B lymphocyte development. Autophagy 2008, 4, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.-H.; Liu, F.; Yang, Z.-L.; Fu, X.-H.; Yang, Z.-H.; Liu, Q.; Wang, L.; Wan, X.-B.; Fan, X.-J. Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am. J. Transl. Res. 2016, 8, 3831–3847. [Google Scholar]
- Oral, O.; Oz-Arslan, D.; Itah, Z.; Naghavi, A.; Deveci, R.; Karacali, S.; Gozuacik, D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012, 17, 810–820. [Google Scholar] [CrossRef]
- Fu, Y.; Hong, L.; Xu, J.; Zhong, G.; Gu, Q.; Gu, Q.; Guan, Y.; Zheng, X.; Dai, Q.; Luo, X.; et al. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy 2019, 15, 295–311. [Google Scholar] [CrossRef] [Green Version]
- Akin, D.; Wang, S.K.; Habibzadegah-Tari, P.; Law, B.; Ostrov, D.; Li, M.; Yin, X.-M.; Kim, J.-S.; Horenstein, N.; Dunn, W.A., Jr. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 2014, 10, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Apel, A.; Herr, I.; Schwarz, H.; Rodemann, H.P.; Mayer, A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008, 68, 1485–1494. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Cheng, F.; Liu, C.X.; Zou, K. Content determination of a steroid saponin from rhizome of Reineckia carnea. J. Huazhong Norm. Univ. 2013, 47, 60–62, 77. (In Chinese) [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, F.-F.; Zhang, J.; Cheng, F.; Zou, K.; Zhang, X.-Q.; Chen, J.-F. ATG 4B Serves a Crucial Role in RCE-4-Induced Inhibition of the Bcl-2–Beclin 1 Complex in Cervical Cancer Ca Ski Cells. Int. J. Mol. Sci. 2021, 22, 12302. https://doi.org/10.3390/ijms222212302
You F-F, Zhang J, Cheng F, Zou K, Zhang X-Q, Chen J-F. ATG 4B Serves a Crucial Role in RCE-4-Induced Inhibition of the Bcl-2–Beclin 1 Complex in Cervical Cancer Ca Ski Cells. International Journal of Molecular Sciences. 2021; 22(22):12302. https://doi.org/10.3390/ijms222212302
Chicago/Turabian StyleYou, Fang-Fang, Jing Zhang, Fan Cheng, Kun Zou, Xue-Qing Zhang, and Jian-Feng Chen. 2021. "ATG 4B Serves a Crucial Role in RCE-4-Induced Inhibition of the Bcl-2–Beclin 1 Complex in Cervical Cancer Ca Ski Cells" International Journal of Molecular Sciences 22, no. 22: 12302. https://doi.org/10.3390/ijms222212302





