Chemoenzymatic Synthesis and Antibody Binding of HIV-1 V1/V2 Glycopeptide-Bacteriophage Qβ Conjugates as a Vaccine Candidate
Abstract
:1. Introduction
2. Results and Discussion
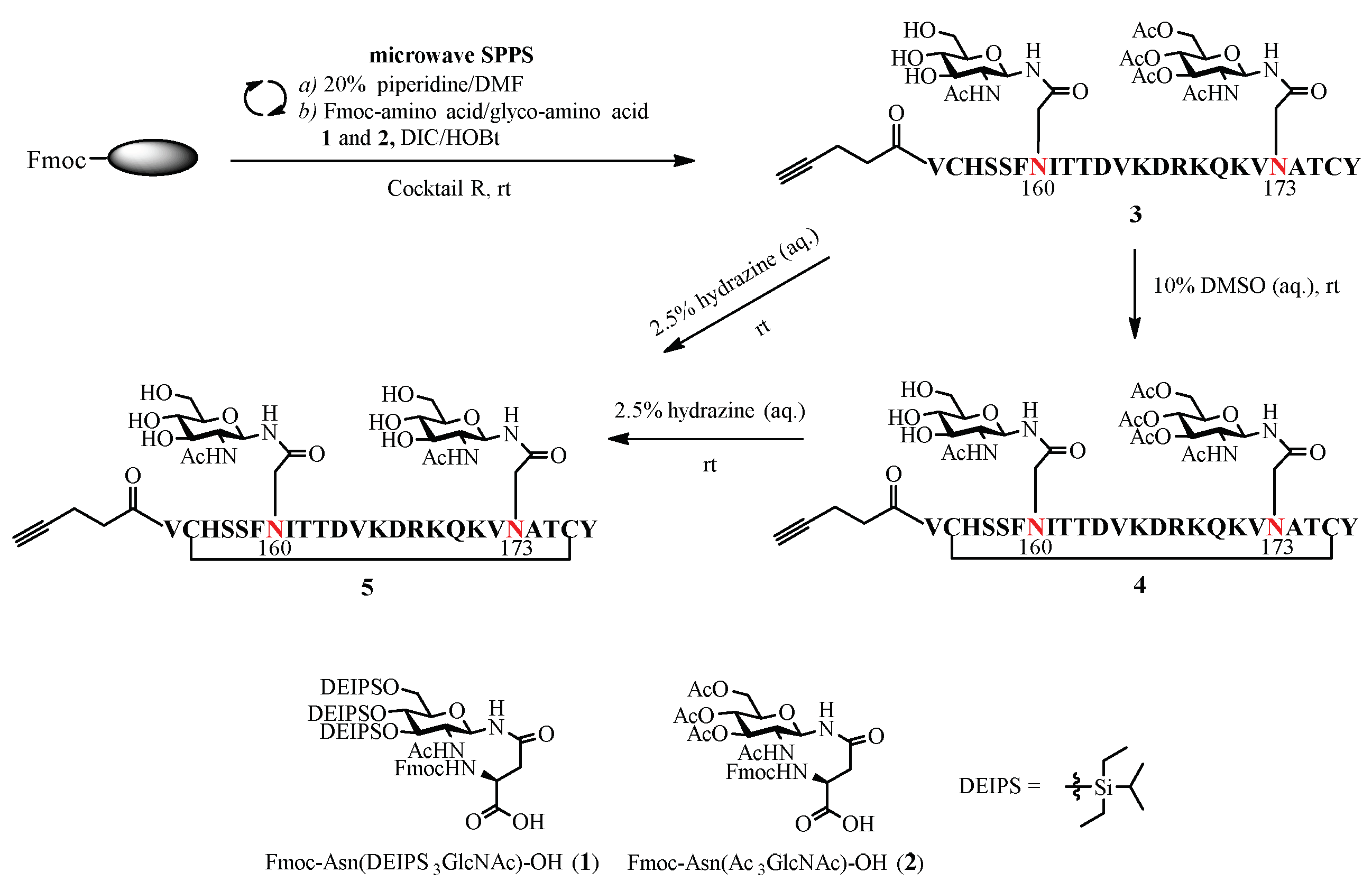
2.1. Chemoenzymatic Synthesis of Alkyne-Tagged HIV-1 V1V2 Glycopeptides
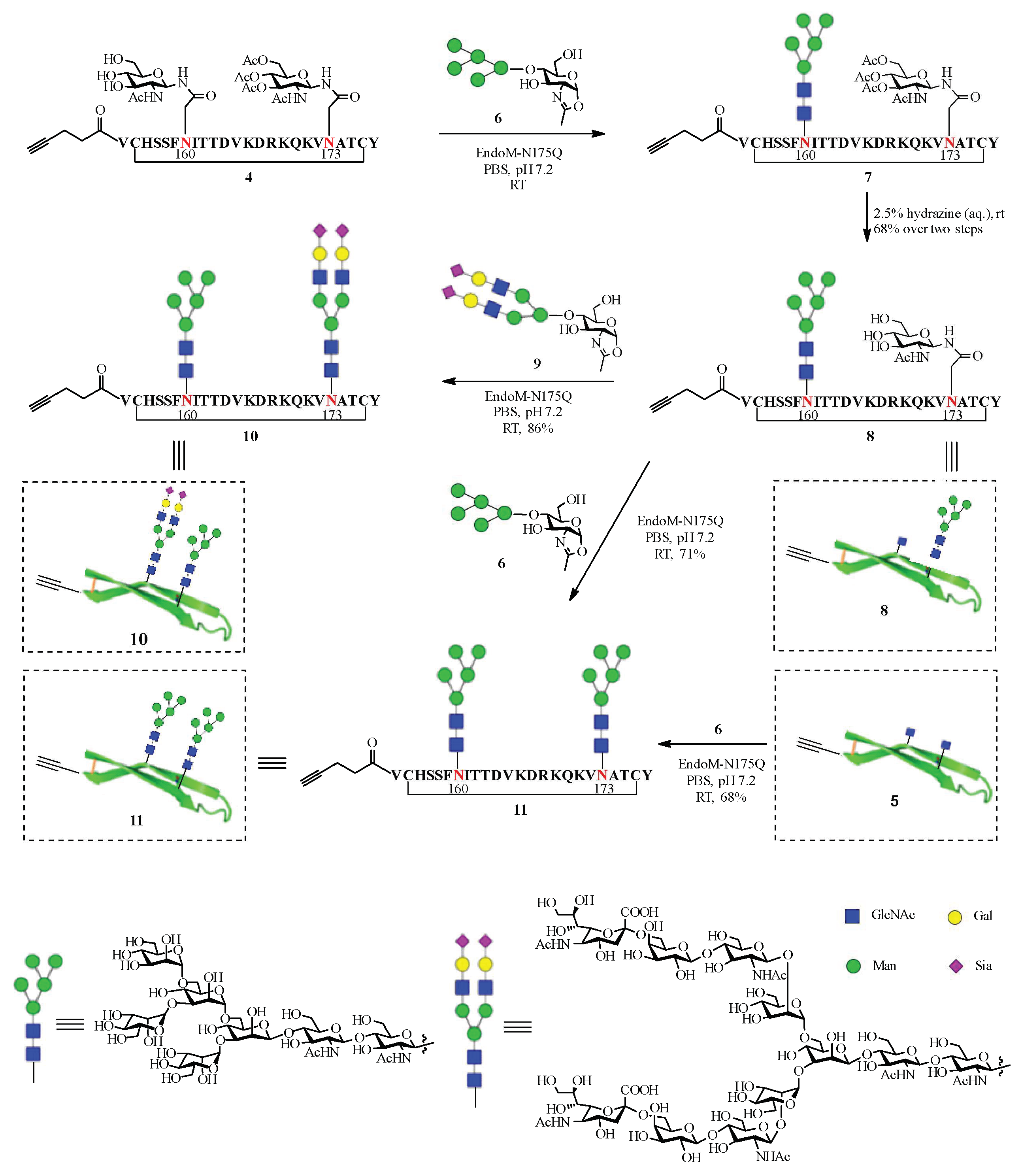
2.2. Chemoenzymatic Synthesis of the HIV-1 V1V2 Glycopeptides
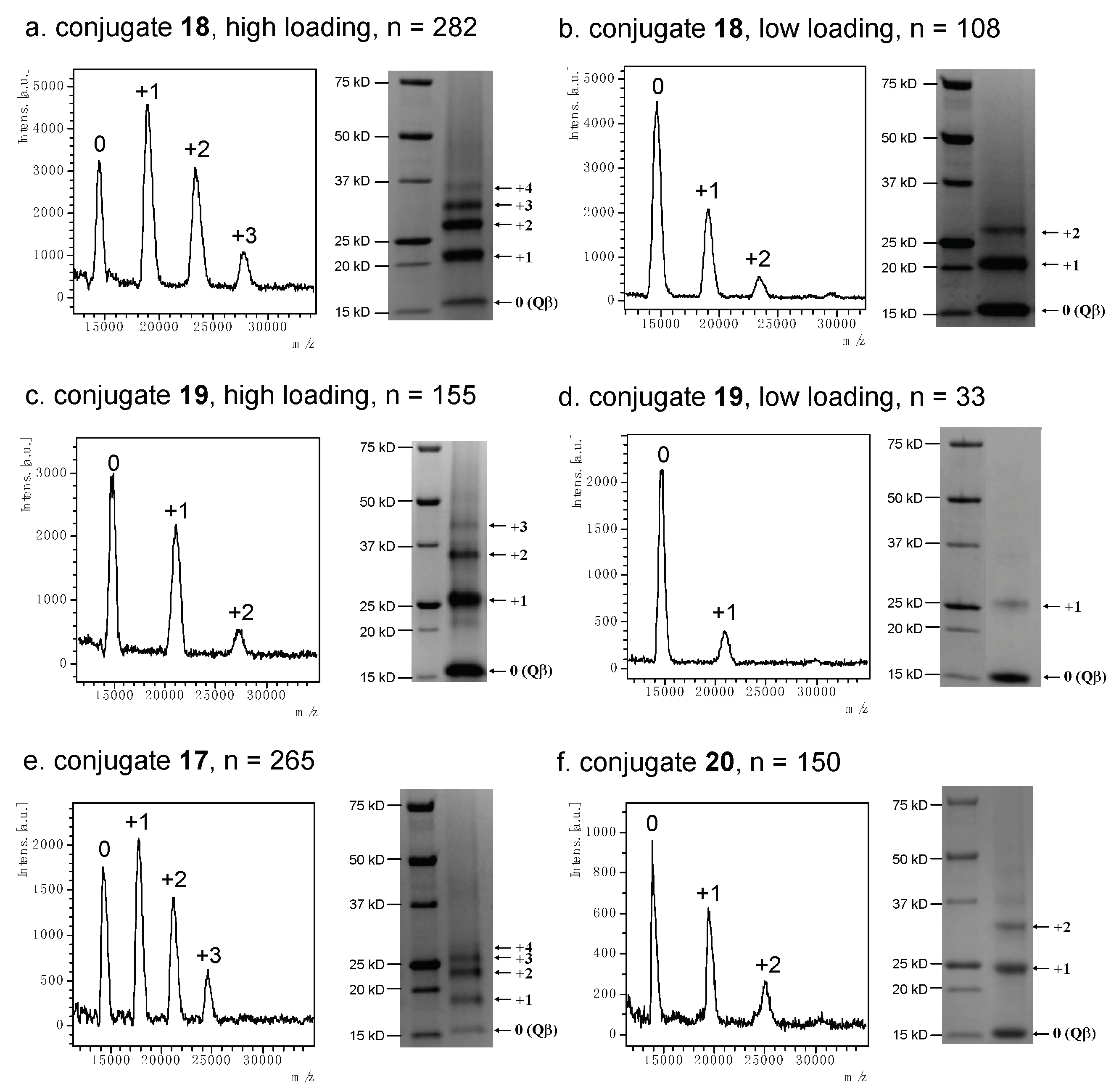
2.3. Synthesis and Characterization of Qβ-V1V2 Glycopeptide Conjugates
2.4. Binding of the Synthetic Conjugates with Antibody PG9
3. Materials and Methods
3.1. General Procedures
3.2. Peptide Synthesis
3.2.1. Synthesis of GlcNAc-Peptide 4 Bearing a GlcNAc at Asn160 and a Ac3GlcNAc at Asn173
3.2.2. Synthesis of GlcNAc-Peptide 5 Bearing a GlcNAc at Asn160 and a GlcNAc at Asn173
3.3. EndoM Mutant-Catalyzed Transglycosylation
3.3.1. Synthesis of Glycopeptide 8 Bearing a Man5GlcNAc2 Glycan at Asn160 and a GlcNAc at Asn173
3.3.2. Synthesis of Glycopeptide 10 Bearing a Man5GlcNAc2 Glycan at Asn160 and a Sialylated Complex-Type Glycan (SCT) at Asn173
3.3.3. Synthesis of Glycopeptide 11 Bearing a Man5GlcNAc2 Glycan at Asn160 and a Man5GlcNAc2 Glycan at Asn173
3.4. Expression and Functionalization of Bacteriophage Qβ Particles
3.4.1. Expression of Wild Type Bacteriophage Qβ
3.4.2. Functionalization of WT Qβ
3.5. Conjugation of V1V2 Glycopeptides to Qβ through CuAAC
3.5.1. General Reaction Conditions for CuAAC
3.5.2. SDS-PAGE Sample Preparation and Analysis
3.5.3. MALDI-TOF MS Sample Preparation and Analysis
3.6. ELISA Analysis of the Binding between PG9 and Glycopeptide-Qβ Conjugate
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nabel, G.J. Challenges and opportunities for development of an AIDS vaccine. Nat. Cell Biol. 2001, 410, 1002–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zolla-Pazner, S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 2004, 4, 199–210. [Google Scholar] [CrossRef]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; De Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.M.; Burton, D.R. Rational antibody-based HIV-1 vaccine design: Current approaches and future directions. Curr. Opin. Immunol. 2010, 22, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Virgin, H.W.; Walker, B.D. Immunology and the elusive AIDS vaccine. Nat. Cell Biol. 2010, 464, 224–231. [Google Scholar] [CrossRef]
- Kim, J.H.; Rerks-Ngarm, S.; Excler, J.-L.; Michael, N.L. HIV vaccines: Lessons learned and the way forward. Curr. Opin. HIV AIDS 2010, 5, 428–434. [Google Scholar] [CrossRef] [Green Version]
- Burton, D.R.; Ahmed, R.; Barouch, D.H.; Butera, S.T.; Crotty, S.; Godzik, A.; Kaufmann, D.E.; McElrath, M.J.; Nussenzweig, M.C.; Pulendran, B.; et al. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe 2012, 12, 396–407. [Google Scholar] [CrossRef] [Green Version]
- Kwong, P.D.; Mascola, J.R.; Nabel, G.J. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: The end of the beginning. Nat. Rev. Immunol. 2013, 13, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Sok, D.; Le, K.M.; Vadnais, M.; Saye-Francisco, K.L.; Jardine, J.G.; Torres, J.L.; Berndsen, Z.T.; Kong, L.; Stanfield, R.; Ruiz, J.; et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nat. Cell Biol. 2017, 548, 108–111. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Crotty, S. HIV vaccinology: 2021 update. Semin. Immunol. 2021, 51, 101470. [Google Scholar] [CrossRef]
- Kwong, P.D.; Mascola, J.R.; Nabel, G.J. The changing face of HIV vaccine research. J. Int. AIDS Soc. 2012, 15, 17407. [Google Scholar] [CrossRef]
- Wei, X.; Decker, J.M.; Wang, S.; Hui, H.; Kappes, J.C.; Wu, X.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Kilby, J.M.; Saag, M.S.; et al. Antibody neutralization and escape by HIV. Nature 2003, 422, 307–312. [Google Scholar] [CrossRef]
- Trkola, A.; Purtscher, M.; Muster, T.; Ballaun, C.; Buchacher, A.; Sullivan, N.; Srinivasan, K.; Sodroski, J.; Moore, J.P.; Katinger, H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type. J. Virol. 1996, 70, 1100–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, R.W.; Venturi, M.; Schiffner, L.; Kalyanaraman, R.; Katinger, H.; Lloyd, K.O.; Kwong, P.D.; Moore, J.P. The Mannose-Dependent Epitope for Neutralizing Antibody 2G12 on Human Immunodeficiency Virus Type 1 Glycoprotein gp. J. Virol. 2002, 76, 7293–7305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, C.N.; Pantophlet, R.; Wormald, M.; Saphire, E.O.; Stanfield, R.; Wilson, I.A.; Katinger, H.; Dwek, R.A.; Rudd, P.M.; Burton, D.R. The Broadly Neutralizing Anti-Human Immunodeficiency Virus Type 1 Antibody 2G12 Recognizes a Cluster of α1→2 Mannose Residues on the Outer Face of gp. J. Virol. 2002, 76, 7306–7321. [Google Scholar] [CrossRef] [Green Version]
- Calarese, D.A.; Scanlan, C.N.; Zwick, M.B.; Deechongkit, S.; Mimura, Y.; Kunert, R.; Zhu, P.; Wormald, M.R.; Stanfield, R.L.; Roux, K.H.; et al. Antibody Domain Exchange Is an Immunological Solution to Carbohydrate Cluster Recognition. Science 2003, 300, 2065–2071. [Google Scholar] [CrossRef]
- Calarese, D.A.; Lee, H.-K.; Huang, C.-Y.; Best, M.D.; Astronomo, R.D.; Stanfield, R.L.; Katinger, H.; Burton, D.R.; Wong, C.-H.; Wilson, I.A. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G. Proc. Natl. Acad. Sci. USA 2005, 102, 13372–13377. [Google Scholar] [CrossRef] [Green Version]
- Walker, L.M.; Phogat, S.K.; Chan-Hui, P.-Y.; Wagner, D.; Phung, P.; Goss, J.L.; Wrin, T.; Simek, M.D.; Fling, S.; Mitcham, J.L.; et al. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science 2009, 326, 285–289. [Google Scholar] [CrossRef] [Green Version]
- Walker, L.M.; Simek, M.D.; Priddy, F.; Gach, J.S.; Wagner, D.; Zwick, M.B.; Phogat, S.K.; Poignard, P.; Burton, D.R. A Limited Number of Antibody Specificities Mediate Broad and Potent Serum Neutralization in Selected HIV-1 Infected Individuals. PLoS Pathog. 2010, 6, e1001028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doores, K.; Burton, D.R. Variable Loop Glycan Dependency of the Broad and Potent HIV-1-Neutralizing Antibodies PG9 and PG. J. Virol. 2010, 84, 10510–10521. [Google Scholar] [CrossRef] [Green Version]
- Bonsignori, M.; Hwang, K.K.; Chen, X.; Tsao, C.Y.; Morris, L.; Gray, E.; Marshall, D.J.; Crump, J.A.; Kapiga, S.H.; Sam, N.E.; et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011, 85, 9998–10009. [Google Scholar] [CrossRef] [Green Version]
- Walker, L.M.; Huber, M.; Doores, K.; Falkowska, E.; Pejchal, R.; Julien, J.-P.; Wang, S.-K.; Ramos, A.; Chan-Hui, P.-Y.; Moyle, M.; et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nat. Cell Biol. 2011, 477, 466–470. [Google Scholar] [CrossRef] [Green Version]
- Pejchal, R.; Doores, K.J.; Walker, L.M.; Khayat, R.; Huang, P.-S.; Wang, S.-K.; Stanfield, R.L.; Julien, J.-P.; Ramos, A.; Crispin, M.; et al. A Potent and Broad Neutralizing Antibody Recognizes and Penetrates the HIV Glycan Shield. Science 2011, 334, 1097–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLellan, J.S.; Pancera, M.; Carrico, C.; Gorman, J.; Julien, J.P.; Khayat, R.; Louder, R.; Pejchal, R.; Sastry, M.; Dai, K.; et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody. PGNature 2011, 480, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.N.; McLellan, J.; Huang, W.; Orwenyo, J.; Burton, D.R.; Koff, W.C.; Kwong, P.D.; Wang, L.-X. Synthetic glycopeptides reveal the glycan specificity of HIV-neutralizing antibodies. Nat. Chem. Biol. 2013, 9, 521–526. [Google Scholar] [CrossRef] [Green Version]
- Orwenyo, J.; Cai, H.; Giddens, J.; Amin, M.N.; Toonstra, C.; Wang, L.-X. Systematic Synthesis and Binding Study of HIV V3 Glycopeptides Reveal the Fine Epitopes of Several Broadly Neutralizing Antibodies. ACS Chem. Biol. 2017, 12, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Orwenyo, J.; Giddens, J.; Yang, Q.; Zhang, R.; LaBranche, C.C.; Montefiori, D.C.; Wang, L.-X. Synthetic Three-Component HIV-1 V3 Glycopeptide Immunogens Induce Glycan-Dependent Antibody Responses. Cell Chem. Biol. 2017, 24, 1513–1522.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Orwenyo, J.; Guenaga, J.; Giddens, J.; Toonstra, C.; Wyatt, R.T.; Wang, L.-X. Synthetic multivalent V3 glycopeptides display enhanced recognition by glycan-dependent HIV-1 broadly neutralizing antibodies. Chem. Commun. 2017, 53, 5453–5456. [Google Scholar] [CrossRef]
- Cai, H.; Zhang, R.; Orwenyo, J.; Giddens, J.; Yang, Q.; Labranche, C.C.; Montefiori, D.C.; Wang, L.-X. Multivalent Antigen Presentation Enhances the Immunogenicity of a Synthetic Three-Component HIV-1 V3 Glycopeptide Vaccine. ACS Central Sci. 2018, 4, 582–589. [Google Scholar] [CrossRef]
- Cai, H.; Zhang, R.-S.; Orwenyo, J.; Giddens, J.P.; Yang, Q.; LaBranche, C.C.; Montefiori, D.C.; Wang, L.-X. Synthetic HIV V3 Glycopeptide Immunogen Carrying a N334 N-Glycan Induces Glycan-Dependent Antibodies with Promiscuous Site Recognition. J. Med. Chem. 2018, 61, 10116–10125. [Google Scholar] [CrossRef]
- Alam, S.M.; Aussedat, B.; Vohra, Y.; Meyerhoff, R.R.; Cale, E.M.; Walkowicz, W.E.; Radakovich, N.A.; Anasti, K.; Armand, L.; Parks, R.; et al. Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci. Transl. Med. 2017, 9, eaai7521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonsignori, M.; Kreider, E.F.; Fera, D.; Meyerhoff, R.R.; Bradley, T.; Wiehe, K.; Alam, S.M.; Aussedat, B.; Walkowicz, W.E.; Hwang, K.-K.; et al. Staged induction of HIV-1 glycan–dependent broadly neutralizing antibodies. Sci. Transl. Med. 2017, 9, eaai7514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Roldao, A.; Mellado, M.C.M.; Castilho, L.R.; Carrondo, M.; Alves, P. Virus-like particles in vaccine development. Expert Rev. Vaccines 2010, 9, 1149–1176. [Google Scholar] [CrossRef]
- Jardine, J.; Julien, J.-P.; Menis, S.; Ota, T.; Kalyuzhniy, O.; McGuire, A.; Sok, D.; Huang, P.-S.; MacPherson, S.; Jones, M.; et al. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science 2013, 340, 711–716. [Google Scholar] [CrossRef] [Green Version]
- McGuire, A.; Hoot, S.; Dreyer, A.M.; Lippy, A.; Stuart, A.; Cohen, K.W.; Jardine, J.; Menis, S.; Scheid, J.F.; West, A.P.; et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 2013, 210, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Kelsoe, G.; Harrison, S.C.; Kepler, T. B-cell–lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012, 30, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Julien, J.-P.; Lee, J.H.; Cupo, A.; Murin, C.D.; Derking, R.; Hoffenberg, S.; Caulfield, M.J.; King, C.R.; Marozsan, A.J.; Klasse, P.J.; et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG. Proc. Natl. Acad. Sci. USA 2013, 110, 4351–4356. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Ye, J.; DeLaitsch, A.T.; Rashidijahanabad, Z.; Lang, S.; Kakeshpour, T.; Zhao, Y.; Ramadan, S.; Saavedra, P.V.; Yuzbasiyan-Gurkan, V.; et al. Chemoenzymatic Synthesis of 9NHAc-GD2 Antigen to Overcome the Hydrolytic Instability of O -Acetylated-GD2 for Anticancer Conjugate Vaccine Development. Angew. Chem. Int. Ed. 2021, 60, 24179–24188. [Google Scholar] [CrossRef]
- Wu, X.; McKay, C.; Pett, C.; Yu, J.; Schorlemer, M.; Ramadan, S.; Lang, S.; Behren, S.; Westerlind, U.; Finn, M.G.; et al. Synthesis and Immunological Evaluation of Disaccharide Bearing MUC-1 Glycopeptide Conjugates with Virus-like Particles. ACS Chem. Biol. 2019, 14, 2176–2184. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Dulaney, S.; McKay, C.; Baniel, C.; Kaczanowska, K.; Ramadan, S.; Finn, M.G.; Huang, X. Chemical Synthesis of GM2 Glycans, Bioconjugation with Bacteriophage Qβ, and the Induction of Anticancer Antibodies. ChemBioChem 2015, 17, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Z.; Chowdhury, S.; McKay, C.; Baniel, C.; Wright, W.S.; Bentley, P.; Kaczanowska, K.; Gildersleeve, J.C.; Finn, M.; BenMohamed, L.; et al. Significant Impact of Immunogen Design on the Diversity of Antibodies Generated by Carbohydrate-Based Anticancer Vaccine. ACS Chem. Biol. 2015, 10, 2364–2372. [Google Scholar] [CrossRef] [Green Version]
- Astronomo, R.D.; Kaltgrad, E.; Udit, A.K.; Wang, S.-K.; Doores, K.; Huang, C.-Y.; Pantophlet, R.; Paulson, J.C.; Wong, C.-H.; Finn, M.; et al. Defining Criteria for Oligomannose Immunogens for HIV Using Icosahedral Virus Capsid Scaffolds. Chem. Biol. 2010, 17, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Toonstra, C.; Amin, M.N.; Wang, L.-X. Site-Selective Chemoenzymatic Glycosylation of an HIV-1 Polypeptide Antigen with Two Distinct N-Glycans via an Orthogonal Protecting Group Strategy. J. Org. Chem. 2016, 81, 6176–6185. [Google Scholar] [CrossRef] [Green Version]
- Umekawa, M.; Huang, W.; Li, B.; Fujita, K.; Ashida, H.; Wang, L.X.; Yamamoto, K. Mutants of Mucor hiemalis endo-beta-N-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J. Biol. Chem. 2008, 283, 4469–4479. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.C.; Timasheff, S.N. The stabilization of proteins by sucrose. J. Biol. Chem. 1981, 256, 7193–7201. [Google Scholar] [CrossRef]
- Aussedat, B.; Vohra, Y.; Park, P.K.; Fernández-Tejada, A.; Alam, S.M.; Dennison, S.M.; Jaeger, F.H.; Anasti, K.; Stewart, S.; Blinn, J.H.; et al. Chemical Synthesis of Highly Congested gp120 V1V2 N-Glycopeptide Antigens for Potential HIV-1-Directed Vaccines. J. Am. Chem. Soc. 2013, 135, 13113–13120. [Google Scholar] [CrossRef] [Green Version]
- Prabhu, S.K.; Li, C.; Zong, G.; Zhang, R.; Wang, L.-X. Comparative studies on the substrate specificity and defucosylation activity of three α-l-fucosidases using synthetic fucosylated glycopeptides and glycoproteins as substrates. Bioorg. Med. Chem. 2021, 42, 116243. [Google Scholar] [CrossRef]
- Zong, G.; Li, C.; Prabhu, S.K.; Zhang, R.; Zhang, X.; Wang, L.X. A facile chemoenzymatic synthesis of SARS-CoV-2 glycopeptides for probing glycosylation functions. Chem. Commun. 2021, 57, 6804–6807. [Google Scholar] [CrossRef]
- Zong, G.; Li, C.; Wang, L.-X. Chemoenzymatic Synthesis of HIV-1 Glycopeptide Antigens. Methods Mol. Biol. 2020, 2103, 249–262. [Google Scholar]
- Fiedler, J.D.; Brown, S.D.; Lau, J.L.; Finn, M.G. RNA-Directed Packaging of Enzymes within Virus-like Particles. Angew. Chem. Int. Ed. 2010, 49, 9648–9651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Z.; Comellas-Aragones, M.; Chowdhury, S.; Bentley, P.; Kaczanowska, K.; Benmohamed, L.; Gildersleeve, J.C.; Finn, M.G.; Huang, X. Boosting immunity to small tumor-associated carbohydrates with bacteriophage qbeta capsids. ACS Chem. Biol. 2013, 8, 1253–1262. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zong, G.; Toonstra, C.; Yang, Q.; Zhang, R.; Wang, L.-X. Chemoenzymatic Synthesis and Antibody Binding of HIV-1 V1/V2 Glycopeptide-Bacteriophage Qβ Conjugates as a Vaccine Candidate. Int. J. Mol. Sci. 2021, 22, 12538. https://doi.org/10.3390/ijms222212538
Zong G, Toonstra C, Yang Q, Zhang R, Wang L-X. Chemoenzymatic Synthesis and Antibody Binding of HIV-1 V1/V2 Glycopeptide-Bacteriophage Qβ Conjugates as a Vaccine Candidate. International Journal of Molecular Sciences. 2021; 22(22):12538. https://doi.org/10.3390/ijms222212538
Chicago/Turabian StyleZong, Guanghui, Christian Toonstra, Qiang Yang, Roushu Zhang, and Lai-Xi Wang. 2021. "Chemoenzymatic Synthesis and Antibody Binding of HIV-1 V1/V2 Glycopeptide-Bacteriophage Qβ Conjugates as a Vaccine Candidate" International Journal of Molecular Sciences 22, no. 22: 12538. https://doi.org/10.3390/ijms222212538





