Structural Determination and Genetic Identification of the O-Antigen from an Escherichia coli Strain, LL004, Representing a Novel Serogroup
Abstract
:1. Introduction
2. Results and Discussion
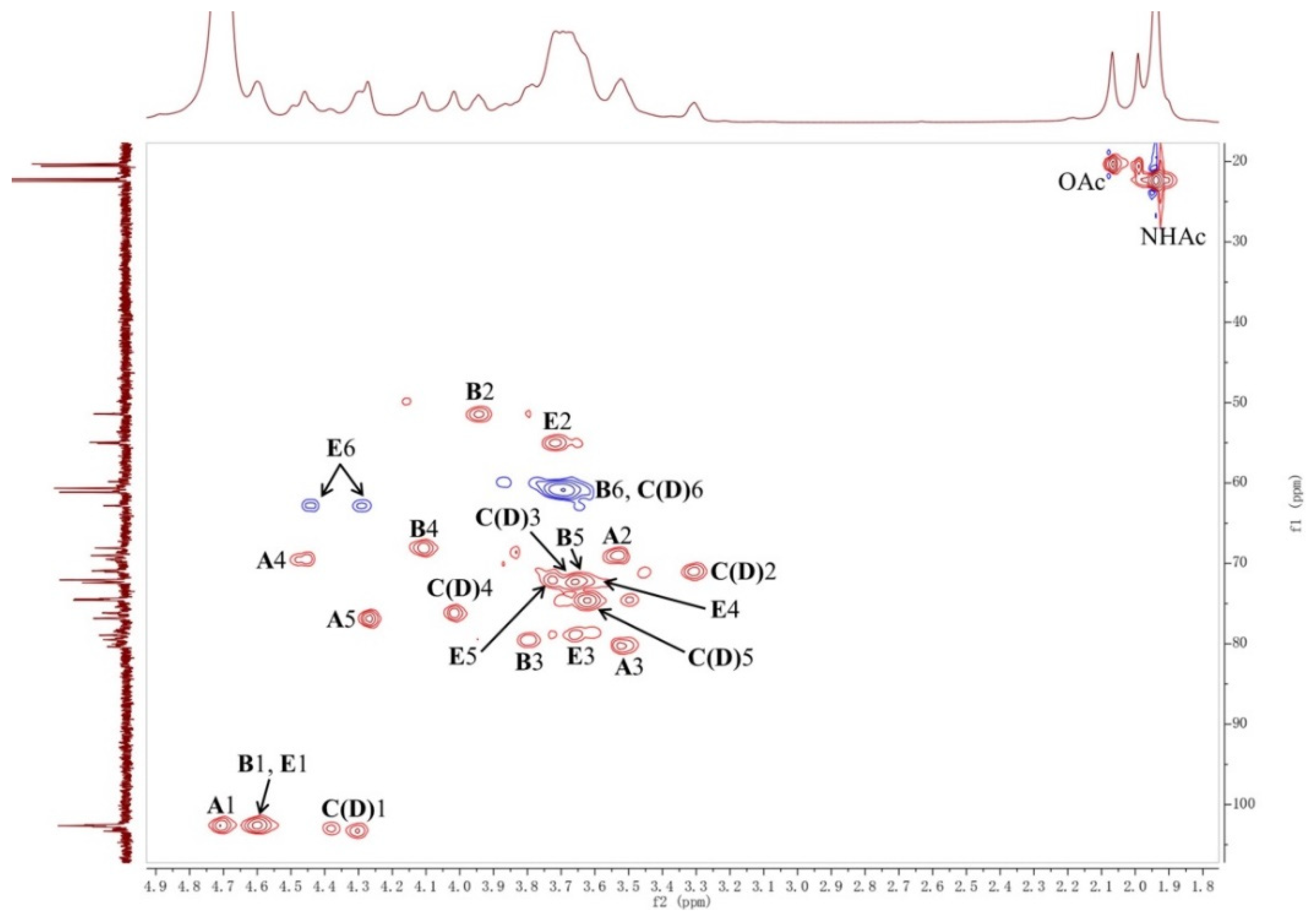
2.1. Structural Analysis of LL004 OAg
2.2. LL004 O-AGC Is Correlated Well to the OAg Structure
2.3. Deletion and Complementation Testing Confirmed the Functionality of the LL004 O-AGC
3. Materials and Methods
3.1. LPS and O-Specific Polysaccharide Extraction
3.2. Monosaccharide Analysis
3.3. NMR Analysis
3.4. Bacterial Strains, Plasmids, and Growth Conditions
3.5. Genome Sequencing and Annotation
3.6. SDS-PAGE Analysis of LPS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Okuda, S.; Sherman, D.J.; Silhavy, T.J.; Ruiz, N.; Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 2016, 14, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Merino, S.; Gonzalez, V.; Tomás, J.M. The first sugar of the repeat units is essential for the Wzy polymerase activity and elongation of the O-antigen lipopolysaccharide. Future Microbiol. 2016, 11, 903–918. [Google Scholar] [CrossRef] [Green Version]
- Dicks, L.; Geldenhuys, J.; Mikkelsen, L.S.; Brandsborg, E.; Marcotte, H. Our gut microbiota: A long walk to homeostasis. Benef. Microbes 2018, 9, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, A.; Iyoda, S.; Kikuchi, T.; Ogura, Y.; Katsura, K.; Ohnishi, M.; Hayashi, T.; Thomson, N.R. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 2015, 22, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.T.; Lam, J.S. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 2014, 60, 697–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenfield, L.K.; Whitfield, C. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 2012, 356, 12–24. [Google Scholar] [CrossRef]
- Chen, M.; Shpirt, A.M.; Guo, X.; Shashkov, A.S.; Zhuang, Y.; Wang, L.; Knirel, Y.A.; Liu, B. Identification serologically, chemically and genetically of two Escherichia coli strains as candidates for new O serogroups. Microbiology 2015, 161, 1790–1796. [Google Scholar] [CrossRef]
- Iguchi, A.; von Mentzer, A.; Kikuchi, T.; Thomson, N.R. An untypeable enterotoxigenic Escherichia coli represents one of the dominant types causing human disease. Microb. Genom. 2017, 3, e000121. [Google Scholar] [CrossRef]
- Lang, C.; Hiller, M.; Konrad, R.; Fruth, A.; Flieger, A. Whole-Genome-Based Public Health Surveillance of Less Common Shiga Toxin-Producing Escherichia coli Serovars and Untypeable Strains Identifies Four Novel O Genotypes. J. Clin. Microbiol. 2019, 57, e00768-19. [Google Scholar] [CrossRef] [Green Version]
- Iguchi, A.; Iyoda, S.; Seto, K.; Morita-Ishihara, T.; Scheutz, F.; Ohnishi, M. Pathogenic E. coli Working Group in Japan. Escherichia coli O-Genotyping PCR: A Comprehensive and Practical Platform for Molecular O Serogrouping. J. Clin. Microbial. 2015, 53, 2427–2432. [Google Scholar]
- Zhao, G.; Perepelov, A.V.; Senchenkova, S.N.; Shashkov, A.S.; Feng, L.; Li, X.; Knirel, Y.A.; Wang, L. Structural relation of the antigenic polysaccharides of Escherichia coli O40, Shigella dysenteriae type 9, and E. coli K47. Carbohydr. Res. 2007, 342, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Perry, M.B.; MacLean, L.L. Structural characterization of the antigenic O-chain of the lipopolysaccharide of Escherichia coli serotype O65. Carbohydr. Res. 1999, 322, 57–66. [Google Scholar] [CrossRef]
- Shashkov, A.S.; Senchenkova, S.N.; Vinogradov, E.V.; Zatonsky, G.V.; Knirel, Y.A.; Literacka, E.; Kaca, W. Full structure of the O-specific polysaccharide of Proteus mirabilis O24 containing 3,4-O-[(S)-1-carboxyethylidene]-d-galactose. Carbohydr. Res. 2000, 329, 453–457. [Google Scholar] [CrossRef]
- Al-Dabbagh, B.; Olatunji, S.; Crouvoisier, M.; El Ghachi, M.; Blanot, D.; Mengin-Lecreulx, D.; Bouhss, A. Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie 2016, 127, 249–257. [Google Scholar] [CrossRef]
- Cunneen, M.M.; Liu, B.; Wang, L.; Reeves, P.R. Biosynthesis of UDP-GlcNAc, UndPP-GlcNAc and UDP-GlcNAcA involves three easily distinguished 4-epimerase enzymes, Gne, Gnu and GnaB. PLoS ONE 2013, 8, e67646. [Google Scholar] [CrossRef]
- Guo, H.; Li, L.; Wang, P.G. Biochemical characterization of UDP-GlcNAc/Glc 4-epimerase from Escherichia coli O86:B7. Biochemistry 2006, 45, 13760–13768. [Google Scholar] [CrossRef]
- Muñoz, R.; López, R.; de Frutos, M.; García, E. First molecular characterization of a uridine diphosphate galacturonate 4-epimerase: An enzyme required for capsular biosynthesis in Streptococcus pneumoniae type 1. Mol. Microbial. 1999, 31, 703–713. [Google Scholar] [CrossRef]
- Liu, B.; Knirel, Y.A.; Feng, L.; Perepelov, A.V.; Senchenkova, S.N.; Wang, Q.; Reeves, P.R.; Wang, L. Structure and genetics of Shigella O antigens. FEMS Microbial. Rev. 2008, 32, 627–653. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.W.; Xia, C.; Li, L.; Guan, W.Y.; Pettit, N.; Zhang, H.C.; Chen, M.; Wang, P.G. Characterization and synthetic application of a novel beta1,3-galactosyltransferase from Escherichia coli O55:H7. Bioorg. Med. Chem. 2009, 17, 4910–4915. [Google Scholar] [CrossRef]
- Hygge Blakeman, K.; Weintraub, A.; Widmalm, G. Structural determination of the O-antigenic polysaccharide from the enterotoxigenic Escherichia coli O147. Eur. J. Biochem. 1998, 251, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Yi, W.; Shao, J.; Lu, Y.; Zhang, W.; Song, J.; Wang, P.G. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz). Appl. Environ. Microbiol. 2005, 71, 7995–8001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robbins, P.W.; Uchida, T. Studies on the chemical basis of the phage conversion of O-antigens in the E-group Salmonellae. Biochemistry 1962, 1, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.Y.; Ren, L.; Liu, W.J.; Sui, Y.; Nong, Q.N.; Xiao, Q.H.; Li, X.Q.; Cao, W. Structural characteristics of a hypoglycemic polysaccharide from Fructus Corni. Carbohydr. Res. 2021, 506, 108358. [Google Scholar] [CrossRef]
- Senchenkova, S.N.; Shashkov, A.S.; Knirel, Y.A.; Esteve, C.; Alcaide, E.; Merino, S.; Tomás, J.M. Structure of a polysaccharide from the lipopolysaccharide of Vibrio vulnificus clinical isolate YJ016 containing 2-acetimidoylamino-2-deoxy-L-galacturonic acid. Carbohydr. Res. 2009, 344, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Feng, F.; Yuan, F.; Su, J.; Cheng, Y.; Wu, H.; Song, K.; Nie, B.; Yu, L.; Zhang, F. Simultaneous determination of 13 carbohydrates using high-performance anion-exchange chromatography coupled with pulsed amperometric detection and mass spectrometry. J. Sep. Sci. 2017, 40, 1843–1854. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Leann, L.; MacLean and Malcolm, B.P. Structural characterization of the serotype O:5 O-polysaccharide antigen of the lipopolysaccharide of Escherichia coli O:5. Biochem. Cell Biol. 1997, 75, 199–205. [Google Scholar]
- Nguyen, T.; Iguchi, A.; Ohata, R.; Kawai, H.; Ooka, T.; Nakajima, H.; Iyoda, S. Distribution of Novel Og Types in Shiga Toxin-Producing Escherichia coli Isolated from Healthy Cattle. J. Clin. Microbial. 2021, 59, e02624-20. [Google Scholar] [CrossRef] [PubMed]






| Residue | Chemical Shifts (ppm) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1-H/1-C | 2-H/2-C | 3-H/3-C | 4-H/4-C | 5-H/5-C | 6-H/6-C | NAc (C=O) | OAc (C=O) | |
| A β-d-GalpA 1JH1,C1 160.3 Hz | 4.71 (3JH,H 6.3 Hz)/102.7 | 3.55 (3JH,H 11.0, 6.3 Hz)/69.1 | 3.52 (3JH,H 11.0 Hz)/80.4 | 4.47 (singlet), 4.50 (singlet)/69.6 | 4.27/76.9 | -/175.0 | - | |
| B β-d-GalpNAc 1JH1,C1 165.2 Hz | 4.60 (3JH,H 8.9 Hz)/102.6 | 3.95 (3JH,H 8.9, 8.9 Hz)/51.4 | 3.80 (3JH,H 8.9 Hz)/79.6 | 4.12 (singlet)/68.1 | 3.63/72.3 | 3.69/60.8 | 1.94/22.4 (174.9) | |
| C(D) β-d-Galp 1JH1,C1 160.2 Hz | 4.38 (3JH,H 6.2 Hz)/103.0, 4.30 (3JH,H 6.4 Hz)/103.3 | 3.31 (3JH,H 9.0, 6.4 Hz)/71.1 | 3.67 (3JH,H 9.0 Hz)/72.3 | 4.02 (singlet)/76.3 | 3.62/74.7 | 3.69/60.8 | - | - |
| E β-d-GlcpNAc6OAc 1JH1,C1 165.2 Hz | 4.61 (3JH,H 11.0 Hz)/102.6 | 3.72 (3JH,H 11.0, 11.0 Hz)/55.1 | 3.64 (3JH,H 11.0 Hz)/79.0 | 3.65/72.5 | 3.73/72.1 | 4.44, 4.31/62.8 | 1.94/22.4 (174.9) | 2.07/20.3 (174.0) |
| Residue | Chemical Shifts (ppm) | ||||||
|---|---|---|---|---|---|---|---|
| 1-H/1-C | 2-H/2-C | 3-H/3-C | 4-H/4-C | 5-H/5-C | 6-H/6-C | NAc (C=O) | |
| A β-d-GalpA | 4.70 (3JH,H 6.6 Hz)/102.6 | 3.51 (3JH,H 10.8, 6.6 Hz)/69.0 | 3.48/80.2 | 4.49 (singlet)/69.7 | 4.21 (singlet)/77.2 | -/175.0 | - |
| B β-d-GalpNAc | 4.57 (3JH,H 9.3 Hz)/02.5 | 3.94 (3JH,H 9.3, 9.3 Hz)/51.4 | 3.79 (3JH,H 9.3 Hz)/79.5 | 4.11 (singlet)/68.0 | 3.62/72.2 | 3.69/60.8 | 1.92/22.2 (175.0) |
| C(D) β-d-Galp | 4.37 (3JH,H 8.6 Hz)/103.0 | 3.29 (3JH,H 8.6, 8.6 Hz)/70.9 | 3.67 (3JH,H 8.6 Hz)/72.4 | 4.00 (singlet)/76.3 | 3.61/74.5 | 3.69/60.8 | - |
| E β-d-GlcpNAc | 4.57 (3JH,H 11.5 Hz)/102.5 | 3.70 (3JH,H 11.5, 8.5 Hz)/54.9 | 3.66/74.8 | 3.61/74.6 | 3.49/74.6 | 3.86, 3.75/59.9 | 1.93/22.2 (175.0) |
| Orf No. | Gene Name | Position of Gene | G+C Content (%) | Similar Protein(s), Strain(s) (Genbank Accession No.) | %Identical/%Similar (Total No. of aa) | Putative Function of Protein |
|---|---|---|---|---|---|---|
| wcaM | 1.1394 | 47.31 | colanic acid biosynthesis protein WcaM [Escherichia coli] (EEU3037011.1) | 99/99 (464) | colanic acid biosynthesis protein | |
| 1 | gnu | 1553.2548 | 49.68 | N-acetyl-alpha-d-glucosaminyl-diphospho-ditrans,octacis-undecaprenol 4-epimerase [Escherichia coli] (WP_113419175.1) | 99/100 (331) | N-acetyl-alpha-d-glucosaminyl-diphospho-ditrans, octacis-undecaprenol 4-epimerase |
| 2 | galF | 2791.3684 | 50.78 | UTP--glucose-1-phosphate uridylyltransferase GalF [Escherichia coli] (WP_113415284.1) | 99/100 (297) | UTP--glucose-1-phosphate uridylyltransferase |
| 3 | wzx | 4151.5272 | 28.6 | oligosaccharide flippase family protein [Aeromonas sp. 1419](WP_216951369.1) | 37/60 (395) | flippase |
| 4 | wzy | 5259.6449 | 28.88 | Wzy [Proteus mirabilis] (AXY99601.1) | 32/49 (394) | polymerase |
| 5 | gtr1 | 6439.7191 | 24.7 | glycosyltransferase family 25 protein [Escherichia coli] (WP_001570044.1) | 100/100 (250) | glycosyltransferase |
| 6 | gtr2 | 7193.8089 | 28.65 | glycosyltransferase family 2 protein [Escherichia coli] (WP_000856120.1) | 100/100 (298) | glycosyltransferase |
| 7 | orf7 | 8100.9362 | 31.03 | hypothetical protein [Escherichia coli] (WP_152696934.1) | 99/100 (420) | hypothetical protein |
| 8 | gtr3 | 9366.10184 | 33.82 | glycosyltransferase [Escherichia coli](WP_001285383.1) | 100/100 (272) | beta-1,3-galactosyltransferase |
| 9 | gnd | 10271.11677 | 49.68 | 6-phosphogluconate dehydrogenase, decarboxylating [Klebsiella pneumoniae IS22] (CDK78112.1) | 99/100 (468) | 6-phosphogluconate dehydrogenase, decarboxylating |
| 10 | ugd | 11926.13092 | 42.93 | UDP-glucose 6-dehydrogenase [Escherichia coli] (BAV90487.1) | 99/100 (397) | UDP-glucose 6-dehydrogenase |
| 11 | gne | complement (13158.14162) | 42.68 | NAD-dependent epimerase [Escherichia coli] (WP_063625168.1) | 99/100 (334) | UDP-N-acetylglucosamine 4-epimerase |
| 12 | wzz | 14571.15548 | 46.72 | chain length determinant protein [Escherichia coli 907357](ESA76081.1) | 99/99 (344) | Chain length determinant protein |
| hisI | complement (15644.16255) | 53.1 | bifunctional phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP diphosphatase HisIE [Escherichia coli](WP_000954885.1) | 100/100 (203) | Phosphoribosyl-AMP cyclohydrolase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Xu, Y.; Qin, C.; Hu, J.; Yin, J.; Guo, X. Structural Determination and Genetic Identification of the O-Antigen from an Escherichia coli Strain, LL004, Representing a Novel Serogroup. Int. J. Mol. Sci. 2021, 22, 12746. https://doi.org/10.3390/ijms222312746
Wang J, Xu Y, Qin C, Hu J, Yin J, Guo X. Structural Determination and Genetic Identification of the O-Antigen from an Escherichia coli Strain, LL004, Representing a Novel Serogroup. International Journal of Molecular Sciences. 2021; 22(23):12746. https://doi.org/10.3390/ijms222312746
Chicago/Turabian StyleWang, Jing, Yujuan Xu, Chunjun Qin, Jing Hu, Jian Yin, and Xi Guo. 2021. "Structural Determination and Genetic Identification of the O-Antigen from an Escherichia coli Strain, LL004, Representing a Novel Serogroup" International Journal of Molecular Sciences 22, no. 23: 12746. https://doi.org/10.3390/ijms222312746





