Single-Strand Annealing in Cancer
Abstract
:1. Introduction
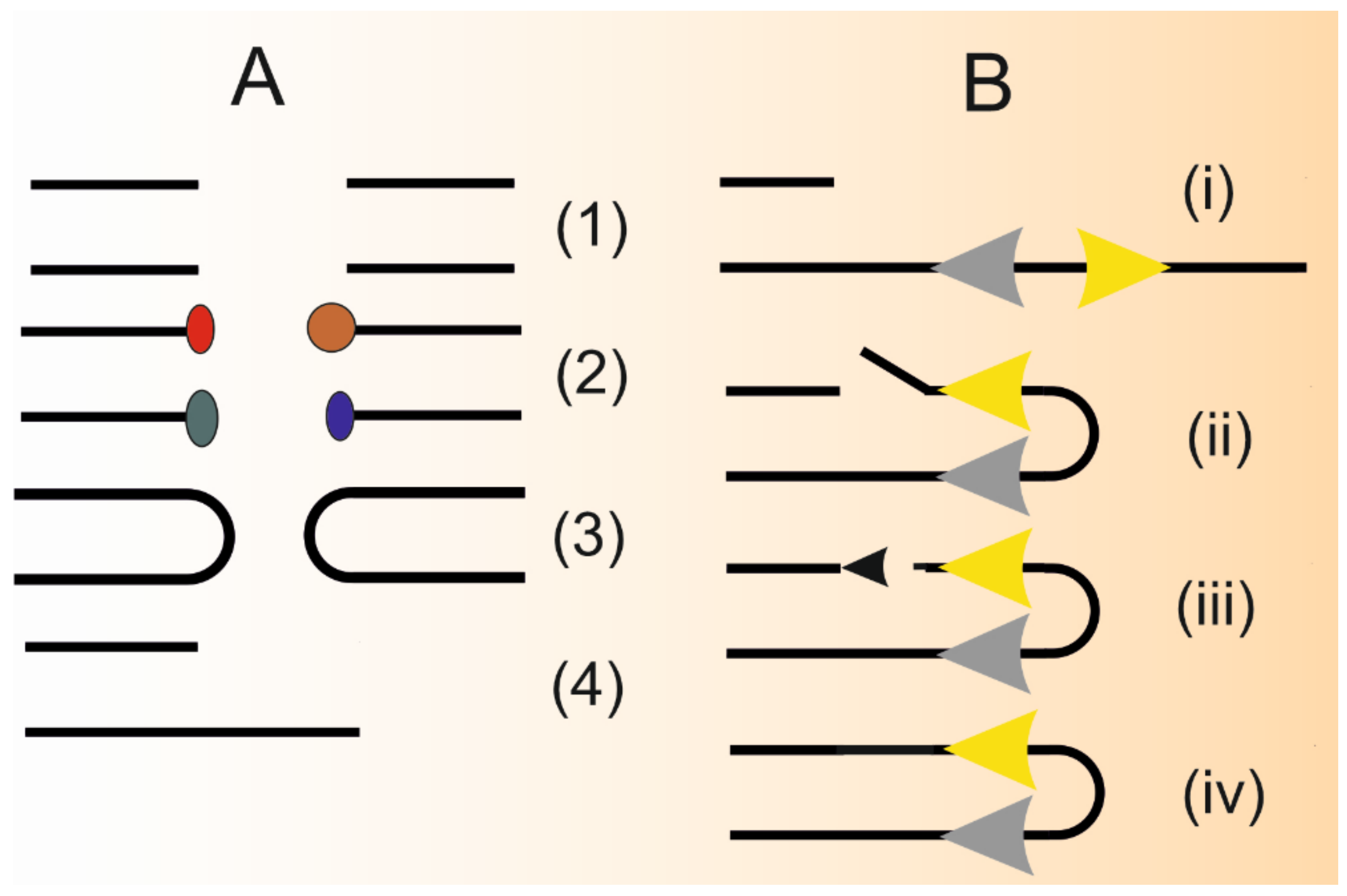
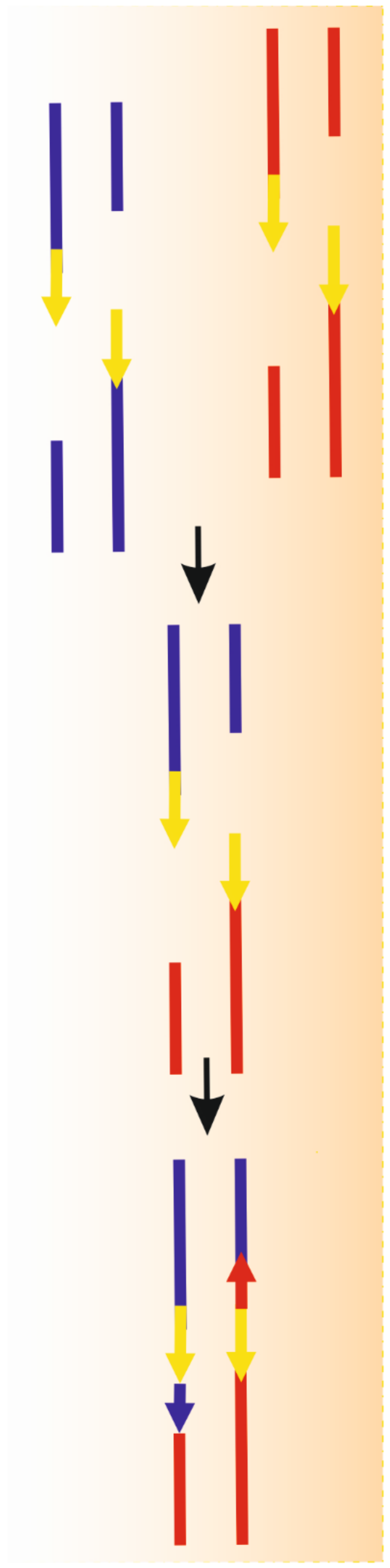
2. Single Strand Annealing–A Distinct Pathway in DNA Double-Strand Break Repair
3. SSA in Genomic Instability and DNA Repair Defects in Cancer
4. SSA in Chromosomal Rearrangements in Cancer
5. SSA in BRCA-Deficient Cells and Cancer Cases
6. SSA in Cancer Clinic
7. SSA in Therapeutic Genome Editing
8. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muramoto, N.; Oda, A.; Tanaka, H.; Nakamura, T.; Kugou, K.; Suda, K.; Kobayashi, A.; Yoneda, S.; Ikeuchi, A.; Sugimoto, H.; et al. Phenotypic diversification by enhanced genome restructuring after induction of multiple DNA double-strand breaks. Nat. Commun. 2018, 9, 1995. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef] [Green Version]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gent, D.C.; Hoeijmakers, J.H.; Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001, 2, 196–206. [Google Scholar] [CrossRef]
- Arnould, C.; Legube, G. The Secret Life of Chromosome Loops upon DNA Double-Strand Break. J. Mol. Biol. 2020, 432, 724–736. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enriquez-Rios, V.; Dumitrache, L.C.; Downing, S.M.; Li, Y.; Brown, E.J.; Russell, H.R.; McKinnon, P.J. DNA-PKcs, ATM, and ATR Interplay Maintains Genome Integrity during Neurogenesis. J. Neurosci. 2017, 37, 893–905. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Front. Genet. 2015, 6, 157. [Google Scholar] [CrossRef] [Green Version]
- Iliakis, G.; Mladenov, E.; Mladenova, V. Necessities in the Processing of DNA Double Strand Breaks and Their Effects on Genomic Instability and Cancer. Cancers 2019, 11, 1671. [Google Scholar] [CrossRef] [Green Version]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin. Cancer Biol. 2016, 37–38, 51–64. [Google Scholar] [CrossRef]
- Seol, J.H.; Shim, E.Y.; Lee, S.E. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. 2018, 809, 81–87. [Google Scholar] [CrossRef]
- Al-Minawi, A.Z.; Saleh-Gohari, N.; Helleday, T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res 2008, 36, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bétermier, M.; Bertrand, P.; Lopez, B.S. Is non-homologous end-joining really an inherently error-prone process? Plos Genet. 2014, 10, e1004086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rass, E.; Grabarz, A.; Plo, I.; Gautier, J.; Bertrand, P.; Lopez, B.S. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009, 16, 819–824. [Google Scholar] [CrossRef] [PubMed]
- So, A.; Le Guen, T.; Lopez, B.S.; Guirouilh-Barbat, J. Genomic rearrangements induced by unscheduled DNA double strand breaks in somatic mammalian cells. FEBS J. 2017, 284, 2324–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huertas, P.; Jackson, S.P. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009, 284, 9558–9565. [Google Scholar] [CrossRef] [Green Version]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Paull, T.T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef]
- Wang, H.; Shi, L.Z.; Wong, C.C.; Han, X.; Hwang, P.Y.; Truong, L.N.; Zhu, Q.; Shao, Z.; Chen, D.J.; Berns, M.W.; et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013, 9, e1003277. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Huang, J. DNA End Resection: Facts and Mechanisms. Genom. Proteom. Bioinform. 2016, 14, 126–130. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Kim, W.; Kloeber, J.A.; Lou, Z. DNA end resection and its role in DNA replication and DSB repair choice in mammalian cells. Exp. Mol. Med. 2020, 52, 1705–1714. [Google Scholar] [CrossRef]
- Sturzenegger, A.; Burdova, K.; Kanagaraj, R.; Levikova, M.; Pinto, C.; Cejka, P.; Janscak, P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 2014, 289, 27314–27326. [Google Scholar] [CrossRef] [Green Version]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef]
- Polato, F.; Callen, E.; Wong, N.; Faryabi, R.; Bunting, S.; Chen, H.T.; Kozak, M.; Kruhlak, M.J.; Reczek, C.R.; Lee, W.H.; et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 2014, 211, 1027–1036. [Google Scholar] [CrossRef]
- Cruz-García, A.; López-Saavedra, A.; Huertas, P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014, 9, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracker, T.H.; Roig, I.; Knobel, P.A.; Marjanović, M. The ATM signaling network in development and disease. Front. Genet. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.J.; Gibb, B.; Kwon, Y.; Sung, P.; Greene, E.C. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res 2017, 45, 749–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, S.K.; Gibb, B.; de Almeida, M.J.; Greene, E.C.; Symington, L.S. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Motycka, T.A.; Bessho, T.; Post, S.M.; Sung, P.; Tomkinson, A.E. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J. Biol. Chem. 2004, 279, 13634–13639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. 2016, 32, 566–575. [Google Scholar] [CrossRef] [Green Version]
- Stefanovie, B.; Hengel, S.R.; Mlcouskova, J.; Prochazkova, J.; Spirek, M.; Nikulenkov, F.; Nemecek, D.; Koch, B.G.; Bain, F.E.; Yu, L.; et al. DSS1 interacts with and stimulates RAD52 to promote the repair of DSBs. Nucleic Acids Res 2020, 48, 694–708. [Google Scholar] [CrossRef]
- Johnson, R.D.; Jasin, M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000, 19, 3398–3407. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.M.; Yanez, D.A.; Stark, J.M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015, 11, e1004943. [Google Scholar] [CrossRef] [Green Version]
- LaRocque, J.R.; Stark, J.M.; Oh, J.; Bojilova, E.; Yusa, K.; Horie, K.; Takeda, J.; Jasin, M. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11971–11976. [Google Scholar] [CrossRef] [Green Version]
- Surtees, J.A.; Alani, E. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 2006, 360, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, N.; Goldfarb, T.; Studamire, B.; Alani, E.; Haber, J.E. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 2004, 101, 9315–9320. [Google Scholar] [CrossRef] [Green Version]
- Tham, K.C.; Kanaar, R.; Lebbink, J.H.G. Mismatch repair and homeologous recombination. DNA Repair 2016, 38, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Abuin, A.; Zhang, H.; Bradley, A. Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol. Cell. Biol. 2000, 20, 149–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, B.; Jasin, M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 2001, 21, 2671–2682. [Google Scholar] [CrossRef] [Green Version]
- Villemure, J.F.; Abaji, C.; Cousineau, I.; Belmaaza, A. MSH2-deficient human cells exhibit a defect in the accurate termination of homology-directed repair of DNA double-strand breaks. Cancer Res. 2003, 63, 3334–3339. [Google Scholar]
- Jahid, S.; Sun, J.; Gelincik, O.; Blecua, P.; Edelmann, W.; Kucherlapati, R.; Zhou, K.; Jasin, M.; Gümüş, Z.H.; Lipkin, S.M. Inhibition of colorectal cancer genomic copy number alterations and chromosomal fragile site tumor suppressor FHIT and WWOX deletions by DNA mismatch repair. Oncotarget 2017, 8, 71574–71586. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhou, X.A.; Zhang, N.; Wang, J. Evolving insights: How DNA repair pathways impact cancer evolution. Cancer Biol. Med. 2020, 17, 805–827. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobachev, K.S.; Gordenin, D.A.; Resnick, M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 2002, 108, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, V.; Mieczkowski, P.A.; Kim, H.M.; Petes, T.D.; Lobachev, K.S. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 2006, 125, 1283–1296. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, S.; Kockler, Z.; Evans, R.; Downing, B.D.; Malkova, A. Single-strand annealing between inverted DNA repeats: Pathway choice, participating proteins, and genome destabilizing consequences. PLoS Genet. 2018, 14, e1007543. [Google Scholar] [CrossRef] [Green Version]
- VanHulle, K.; Lemoine, F.J.; Narayanan, V.; Downing, B.; Hull, K.; McCullough, C.; Bellinger, M.; Lobachev, K.; Petes, T.D.; Malkova, A. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 2007, 27, 2601–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoshani, O.; Brunner, S.F.; Yaeger, R.; Ly, P.; Nechemia-Arbely, Y.; Kim, D.H.; Fang, R.; Castillon, G.A.; Yu, M.; Li, J.S.Z.; et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 2020. [Google Scholar] [CrossRef]
- Galanos, P.; Vougas, K.; Walter, D.; Polyzos, A.; Maya-Mendoza, A.; Haagensen, E.J.; Kokkalis, A.; Roumelioti, F.M.; Gagos, S.; Tzetis, M.; et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016, 18, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Galanos, P.; Pappas, G.; Polyzos, A.; Kotsinas, A.; Svolaki, I.; Giakoumakis, N.N.; Glytsou, C.; Pateras, I.S.; Swain, U.; Souliotis, V.L.; et al. Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol. 2018, 19, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhl, M.; Csernok, A.; Aydin, S.; Kreienberg, R.; Wiesmüller, L.; Gatz, S.A. Role of SIRT1 in homologous recombination. DNA Repair 2010, 9, 383–393. [Google Scholar] [CrossRef]
- Sample, K.M. DNA repair gene expression is associated with differential prognosis between HPV16 and HPV18 positive cervical cancer patients following radiation therapy. Sci. Rep. 2020, 10, 2774. [Google Scholar] [CrossRef] [Green Version]
- Pal, J.; Nanjappa, P.; Kumar, S.; Shi, J.; Buon, L.; Munshi, N.C.; Shammas, M.A. Impact of RAD51C-mediated Homologous Recombination on Genomic Integrity in Barrett’s Adenocarcinoma Cells. J. Gastroenterol. Hepatol. Res. 2017, 6, 2286–2295. [Google Scholar] [CrossRef] [Green Version]
- Goubert, C.; Thomas, J.; Payer, L.M.; Kidd, J.M.; Feusier, J.; Watkins, W.S.; Burns, K.H.; Jorde, L.B.; Feschotte, C. TypeTE: A tool to genotype mobile element insertions from whole genome resequencing data. Nucleic Acids Res 2020, 48, e36. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Bernhardy, A.J.; Nacson, J.; Krais, J.J.; Tan, Y.F.; Nicolas, E.; Radke, M.R.; Handorf, E.; Llop-Guevara, A.; Balmaña, J.; et al. BRCA1 intronic Alu elements drive gene rearrangements and PARP inhibitor resistance. Nat. Commun. 2019, 10, 5661. [Google Scholar] [CrossRef] [Green Version]
- Mehdipour, P.; Marhon, S.A.; Ettayebi, I.; Chakravarthy, A.; Hosseini, A.; Wang, Y.; de Castro, F.A.; Loo Yau, H.; Ishak, C.; Abelson, S.; et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 2020, 588, 169–173. [Google Scholar] [CrossRef]
- Wilch, E.S.; Morton, C.C. Historical and Clinical Perspectives on Chromosomal Translocations. Adv. Exp. Med. Biol. 2018, 1044, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Deininger, M.W.; Shah, N.P.; Altman, J.K.; Berman, E.; Bhatia, R.; Bhatnagar, B.; DeAngelo, D.J.; Gotlib, J.; Hobbs, G.; Maness, L.; et al. Chronic Myeloid Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 1385–1415. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Deininger, M.W. Declaration of Bcr-Abl1 independence. Leukemia 2020, 34, 2827–2836. [Google Scholar] [CrossRef] [PubMed]
- Oehler, V.G. First-generation vs second-generation tyrosine kinase inhibitors: Which is best at diagnosis of chronic phase chronic myeloid leukemia? Hematol. Am. Soc. Hematol. Educ. Program 2020, 2020, 228–236. [Google Scholar] [CrossRef]
- Salles, D.; Mencalha, A.L.; Ireno, I.C.; Wiesmüller, L.; Abdelhay, E. BCR-ABL stimulates mutagenic homologous DNA double-strand break repair via the DNA-end-processing factor CtIP. Carcinogenesis 2011, 32, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Kolomietz, E.; Al-Maghrabi, J.; Brennan, S.; Karaskova, J.; Minkin, S.; Lipton, J.; Squire, J.A. Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood 2001, 97, 3581–3588. [Google Scholar] [CrossRef] [Green Version]
- Cramer, K.; Nieborowska-Skorska, M.; Koptyra, M.; Slupianek, A.; Penserga, E.T.; Eaves, C.J.; Aulitzky, W.; Skorski, T. BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res. 2008, 68, 6884–6888. [Google Scholar] [CrossRef] [Green Version]
- Slupianek, A.; Poplawski, T.; Jozwiakowski, S.K.; Cramer, K.; Pytel, D.; Stoczynska, E.; Nowicki, M.O.; Blasiak, J.; Skorski, T. BCR/ABL stimulates WRN to promote survival and genomic instability. Cancer Res. 2011, 71, 842–851. [Google Scholar] [CrossRef] [Green Version]
- Startek, M.; Szafranski, P.; Gambin, T.; Campbell, I.M.; Hixson, P.; Shaw, C.A.; Stankiewicz, P.; Gambin, A. Genome-wide analyses of LINE-LINE-mediated nonallelic homologous recombination. Nucleic Acids Res 2015, 43, 2188–2198. [Google Scholar] [CrossRef] [Green Version]
- Elliott, B.; Richardson, C.; Jasin, M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol. Cell 2005, 17, 885–894. [Google Scholar] [CrossRef]
- Morales, M.E.; White, T.B.; Streva, V.A.; DeFreece, C.B.; Hedges, D.J.; Deininger, P.L. The contribution of alu elements to mutagenic DNA double-strand break repair. PLoS Genet. 2015, 11, e1005016. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Lu, H.; Wang, H.; Li, S.; Truong, L.; Li, J.; Liu, S.; Xiang, R.; Wu, X. Break-induced replication plays a prominent role in long-range repeat-mediated deletion. Embo J. 2019, 38, e101751. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, C.M.; Ho, B.N.; Singh, A.T.K. The Evolution, Functions and Applications of the Breast Cancer Genes BRCA1 and BRCA2. Cancer Genomics Proteom. 2017, 14, 293–298. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R. Hereditary breast and ovarian cancer (HBOC): Review of its molecular characteristics, screening, treatment, and prognosis. Breast Cancer 2020. [Google Scholar] [CrossRef]
- Yoshida, K.; Miki, Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004, 95, 866–871. [Google Scholar] [CrossRef]
- Towler, W.I.; Zhang, J.; Ransburgh, D.J.; Toland, A.E.; Ishioka, C.; Chiba, N.; Parvin, J.D. Analysis of BRCA1 variants in double-strand break repair by homologous recombination and single-strand annealing. Hum. Mutat. 2013, 34, 439–445. [Google Scholar] [CrossRef] [Green Version]
- Saha, J.; Davis, A.J. Unsolved mystery: The role of BRCA1 in DNA end-joining. J. Radiat. Res. 2016, 57 (Suppl. S1), i18–i24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogueira, A.; Fernandes, M.; Catarino, R.; Medeiros, R. RAD52 Functions in Homologous Recombination and Its Importance on Genomic Integrity Maintenance and Cancer Therapy. Cancers 2019, 11, 1622. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Goyal, N.; Sullivan, K.; Hanamshet, K.; Patel, M.; Mazina, O.M.; Wang, C.X.; An, W.F.; Spoonamore, J.; Metkar, S.; et al. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res 2016, 44, 4189–4199. [Google Scholar] [CrossRef]
- Hodgson, A.; Turashvili, G. Pathology of Hereditary Breast and Ovarian Cancer. Front. Oncol. 2020, 10, 531790. [Google Scholar] [CrossRef]
- Pietrasik, S.; Zajac, G.; Morawiec, J.; Soszynski, M.; Fila, M.; Blasiak, J. Interplay between BRCA1 and GADD45A and Its Potential for Nucleotide Excision Repair in Breast Cancer Pathogenesis. Int. J. Mol. Sci. 2020, 21, 870. [Google Scholar] [CrossRef] [Green Version]
- Sullivan-Reed, K.; Bolton-Gillespie, E.; Dasgupta, Y.; Langer, S.; Siciliano, M.; Nieborowska-Skorska, M.; Hanamshet, K.; Belyaeva, E.A.; Bernhardy, A.J.; Lee, J.; et al. Simultaneous Targeting of PARP1 and RAD52 Triggers Dual Synthetic Lethality in BRCA-Deficient Tumor Cells. Cell Rep. 2018, 23, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Compagnone, M.; Lavorgna, S.; Angelini, D.F.; Cencioni, M.T.; Piras, E.; Panetta, P.; Ottone, T.; Dolci, S.; Venditti, A.; et al. BRCA1, PARP1 and γH2AX in acute myeloid leukemia: Role as biomarkers of response to the PARP inhibitor olaparib. Biochim. Biophys. Acta 2015, 1852, 462–472. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Lin, Y.; Luo, Y.; Yang, Y.; Long, B.; Fang, Z.; Liu, L.; Zhang, J.; Zhang, X. RAD52 aptamer regulates DNA damage repair and STAT3 in BRCA1/BRCA2-deficient human acute myeloid leukemia. Oncol. Rep. 2020, 44, 1455–1466. [Google Scholar] [CrossRef]
- Adamson, A.W.; Ding, Y.C.; Mendez-Dorantes, C.; Bailis, A.M.; Stark, J.M.; Neuhausen, S.L. The RAD52 S346X variant reduces risk of developing breast cancer in carriers of pathogenic germline BRCA2 mutations. Mol. Oncol. 2020, 14, 1124–1133. [Google Scholar] [CrossRef]
- Gao, D.; Zhu, B.; Cao, X.; Zhang, M.; Wang, X. Roles of NIPBL in maintenance of genome stability. Semin. Cell Dev. Biol. 2019, 90, 181–186. [Google Scholar] [CrossRef]
- Muñoz, M.C.; Laulier, C.; Gunn, A.; Cheng, A.; Robbiani, D.F.; Nussenzweig, A.; Stark, J.M. RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J. Biol. Chem. 2012, 287, 40618–40628. [Google Scholar] [CrossRef] [Green Version]
- Meghani, K.; Fuchs, W.; Detappe, A.; Drané, P.; Gogola, E.; Rottenberg, S.; Jonkers, J.; Matulonis, U.; Swisher, E.M.; Konstantinopoulos, P.A.; et al. Multifaceted Impact of MicroRNA 493-5p on Genome-Stabilizing Pathways Induces Platinum and PARP Inhibitor Resistance in BRCA2-Mutated Carcinomas. Cell Rep. 2018, 23, 100–111. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Jasin, M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 2017, 8, 525. [Google Scholar] [CrossRef] [PubMed]
- Obermeier, K.; Sachsenweger, J.; Friedl, T.W.; Pospiech, H.; Winqvist, R.; Wiesmüller, L. Heterozygous PALB2 c.1592delT mutation channels DNA double-strand break repair into error-prone pathways in breast cancer patients. Oncogene, 2016; 35, 3796–3806. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Liu, L.; Xie, N.; Lu, J.; Liu, Z.; Tang, Y.; Wang, Y.; Yang, J.; Ouyang, Q. Germline PALB2 Mutations in Cancers and Its Distinction From Somatic PALB2 Mutations in Breast Cancers. Front. Genet. 2020, 11, 829. [Google Scholar] [CrossRef] [PubMed]
- Ochs, F.; Somyajit, K.; Altmeyer, M.; Rask, M.B.; Lukas, J.; Lukas, C. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016, 23, 714–721. [Google Scholar] [CrossRef]
- Hanamshet, K.; Mazin, A.V. The function of RAD52 N-terminal domain is essential for viability of BRCA-deficient cells. Nucleic Acids Res 2020, 48, 12778–12791. [Google Scholar] [CrossRef] [PubMed]
- Stark, J.M.; Pierce, A.J.; Oh, J.; Pastink, A.; Jasin, M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004, 24, 9305–9316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deniz, M.; Romashova, T.; Kostezka, S.; Faul, A.; Gundelach, T.; Moreno-Villanueva, M.; Janni, W.; Friedl, T.W.P.; Wiesmüller, L. Increased single-strand annealing rather than non-homologous end-joining predicts hereditary ovarian carcinoma. Oncotarget 2017, 8, 98660–98676. [Google Scholar] [CrossRef] [Green Version]
- Benitez, A.; Liu, W.; Palovcak, A.; Wang, G.; Moon, J.; An, K.; Kim, A.; Zheng, K.; Zhang, Y.; Bai, F.; et al. FANCA Promotes DNA Double-Strand Break Repair by Catalyzing Single-Strand Annealing and Strand Exchange. Mol. Cell 2018, 71, 621–628.e624. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, G.; Palovcak, A.; Li, Y.; Hao, S.; Liu, Z.J.; Landgraf, R.; Yuan, F.; Zhang, Y. Impeding the single-strand annealing pathway of DNA double-strand break repair by withaferin A-mediated FANCA degradation. DNA Repair 2019, 77, 10–17. [Google Scholar] [CrossRef]
- Blasiak, J. DNA-Damaging Anticancer Drugs—A Perspective for DNA Repair- Oriented Therapy. Curr. Med. Chem. 2017, 24, 1488–1503. [Google Scholar] [CrossRef]
- Pancheri, E.; Guglielmi, V.; Wilczynski, G.M.; Malatesta, M.; Tonin, P.; Tomelleri, G.; Nowis, D.; Vattemi, G. Non-Hematologic Toxicity of Bortezomib in Multiple Myeloma: The Neuromuscular and Cardiovascular Adverse Effects. Cancers 2020, 12, 2540. [Google Scholar] [CrossRef]
- Jacquemont, C.; Simon, J.A.; D’Andrea, A.D.; Taniguchi, T. Non-specific chemical inhibition of the Fanconi anemia pathway sensitizes cancer cells to cisplatin. Mol. Cancer 2012, 11, 26. [Google Scholar] [CrossRef] [Green Version]
- Keimling, M.; Deniz, M.; Varga, D.; Stahl, A.; Schrezenmeier, H.; Kreienberg, R.; Hoffmann, I.; König, J.; Wiesmüller, L. The power of DNA double-strand break (DSB) repair testing to predict breast cancer susceptibility. FASEB J. 2012, 26, 2094–2104. [Google Scholar] [CrossRef]
- Daisy, P.S.; Shreyas, K.S.; Anitha, T.S. Will CRISPR-Cas9 Have Cards to Play Against Cancer? An Update on its Applications. Mol. Biotechnol. 2021, 1–16. [Google Scholar] [CrossRef]
- Ghaemi, A.; Bagheri, E.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. CRISPR-cas9 genome editing delivery systems for targeted cancer therapy. Life Sci. 2020, 267, 118969. [Google Scholar] [CrossRef]
- Liu, Q. World-First Phase I Clinical Trial for CRISPR-Cas9 PD-1-Edited T-Cells in Advanced Nonsmall Cell Lung Cancer. Glob. Med. Genet. 2020, 7, 73–74. [Google Scholar] [CrossRef]
- Van der Weyden, L.; Jonkers, J.; Adams, D.J. The use of CRISPR/Cas9-based gene editing strategies to explore cancer gene function in mice. Curr. Opin. Genet. Dev. 2021, 66, 57–62. [Google Scholar] [CrossRef]
- Kim, E.; Hart, T. Improved analysis of CRISPR fitness screens and reduced off-target effects with the BAGEL2 gene essentiality classifier. Genome Med. 2021, 13, 2. [Google Scholar] [CrossRef]
- Yokoyama, M.; Matsuzawa, T.; Yoshikawa, T.; Nunomiya, A.; Yamaguchi, Y.; Yanai, K. Heparan sulfate controls skeletal muscle differentiation and motor functions. Biochim. Biophys. Acta. Gen. Subj. 2020, 1864, 129707. [Google Scholar] [CrossRef]
- Hazelbaker, D.Z.; Beccard, A.; Angelini, G.; Mazzucato, P.; Messana, A.; Lam, D.; Eggan, K.; Barrett, L.E. A multiplexed gRNA piggyBac transposon system facilitates efficient induction of CRISPRi and CRISPRa in human pluripotent stem cells. Sci. Rep. 2020, 10, 635. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Bai, Y.; Cheng, X.; Kalds, P.G.T.; Sun, B.; Wu, Y.; Lv, H.; Xu, K.; Zhang, Z. Efficient SSA-mediated precise genome editing using CRISPR/Cas9. FEBS J. 2018, 285, 3362–3375. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Xu, K.; Liu, Z.; Shen, J.; Han, F.; Chen, Z.; Zhang, Z. Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell. Mol. Life Sci. 2015, 72, 2763–2772. [Google Scholar] [CrossRef]
- Jinqing, W.; Gui, M.; Zhiguo, L.; Yaosheng, C.; Peiqing, C.; Zuyong, H. Improving gene targeting efficiency on pig IGF2 mediated by ZFNs and CRISPR/Cas9 by using SSA reporter system. Yi Chuan = Hered. 2015, 37, 55–62. [Google Scholar] [CrossRef]
- Zhang, W.W.; Matlashewski, G. Single-Strand Annealing Plays a Major Role in Double-Strand DNA Break Repair following CRISPR-Cas9 Cleavage in Leishmania. mSphere 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Brunet, E.; Jasin, M. Induction of Chromosomal Translocations with CRISPR-Cas9 and Other Nucleases: Understanding the Repair Mechanisms That Give Rise to Translocations. Adv. Exp. Med. Biol. 2018, 1044, 15–25. [Google Scholar] [CrossRef]
- Zan, H.; Tat, C.; Qiu, Z.; Taylor, J.R.; Guerrero, J.A.; Shen, T.; Casali, P. Rad52 competes with Ku70/Ku86 for binding to S-region DSB ends to modulate antibody class-switch DNA recombination. Nat. Commun. 2017, 8, 14244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, R.L.; Wijnhoven, P.W.G.; Ramos-Montoya, A.; Wilson, Z.; Illuzzi, G.; Falenta, K.; Jones, G.N.; James, N.; Chabbert, C.D.; Stott, J.; et al. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene 2020, 39, 4869–4883. [Google Scholar] [CrossRef]
- Yu, W.; Lescale, C.; Babin, L.; Bedora-Faure, M.; Lenden-Hasse, H.; Baron, L.; Demangel, C.; Yelamos, J.; Brunet, E.; Deriano, L. Repair of G1 induced DNA double-strand breaks in S-G2/M by alternative NHEJ. Nat. Commun. 2020, 11, 5239. [Google Scholar] [CrossRef]
- Hendrickson, E.A. RAD52: Viral Friend or Foe? Cancers 2020, 12, 399. [Google Scholar] [CrossRef] [Green Version]
- Pawlyn, C.; Loehr, A.; Ashby, C.; Tytarenko, R.; Deshpande, S.; Sun, J.; Fedorchak, K.; Mughal, T.; Davies, F.E.; Walker, B.A.; et al. Loss of heterozygosity as a marker of homologous repair deficiency in multiple myeloma: A role for PARP inhibition? Leukemia 2018, 32, 1561–1566. [Google Scholar] [CrossRef] [Green Version]
- Sakofsky, C.J.; Malkova, A. Break induced replication in eukaryotes: Mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 395–413. [Google Scholar] [CrossRef]
- Cody, J.D. The Consequences of Abnormal Gene Dosage: Lessons from Chromosome 18. Trends Genet. 2020, 36, 764–776. [Google Scholar] [CrossRef]
- Taira, K.; Kaneto, S.; Nakano, K.; Watanabe, S.; Takahashi, E.; Arimoto, S.; Okamoto, K.; Schaaper, R.M.; Negishi, K.; Negishi, T. Distinct pathways for repairing mutagenic lesions induced by methylating and ethylating agents. Mutagenesis 2013, 28, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Jalan, M.; Olsen, K.S.; Powell, S.N. Emerging Roles of RAD52 in Genome Maintenance. Cancers 2019, 11, 1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meers, C.; Keskin, H.; Banyai, G.; Mazina, O.; Yang, T.; Gombolay, A.L.; Mukherjee, K.; Kaparos, E.I.; Newnam, G.; Mazin, A.; et al. Genetic Characterization of Three Distinct Mechanisms Supporting RNA-Driven DNA Repair and Modification Reveals Major Role of DNA Polymerase ζ. Mol. Cell 2020, 79, 1037–1050.e1035. [Google Scholar] [CrossRef]
- Welty, S.; Teng, Y.; Liang, Z.; Zhao, W.; Sanders, L.H.; Greenamyre, J.T.; Rubio, M.E.; Thathiah, A.; Kodali, R.; Wetzel, R.; et al. RAD52 is required for RNA-templated recombination repair in post-mitotic neurons. J. Biol. Chem. 2018, 293, 1353–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blasiak, J. Single-Strand Annealing in Cancer. Int. J. Mol. Sci. 2021, 22, 2167. https://doi.org/10.3390/ijms22042167
Blasiak J. Single-Strand Annealing in Cancer. International Journal of Molecular Sciences. 2021; 22(4):2167. https://doi.org/10.3390/ijms22042167
Chicago/Turabian StyleBlasiak, Janusz. 2021. "Single-Strand Annealing in Cancer" International Journal of Molecular Sciences 22, no. 4: 2167. https://doi.org/10.3390/ijms22042167





