Effects of Structural Isomers of Spermine on the Higher-Order Structure of DNA and Gene Expression
Abstract
:1. Introduction
2. Results and Discussion
2.1. Change in The Higher-Order Structure of DNA under Single Molecule Observation with Fluorescence Microscopy (FM)
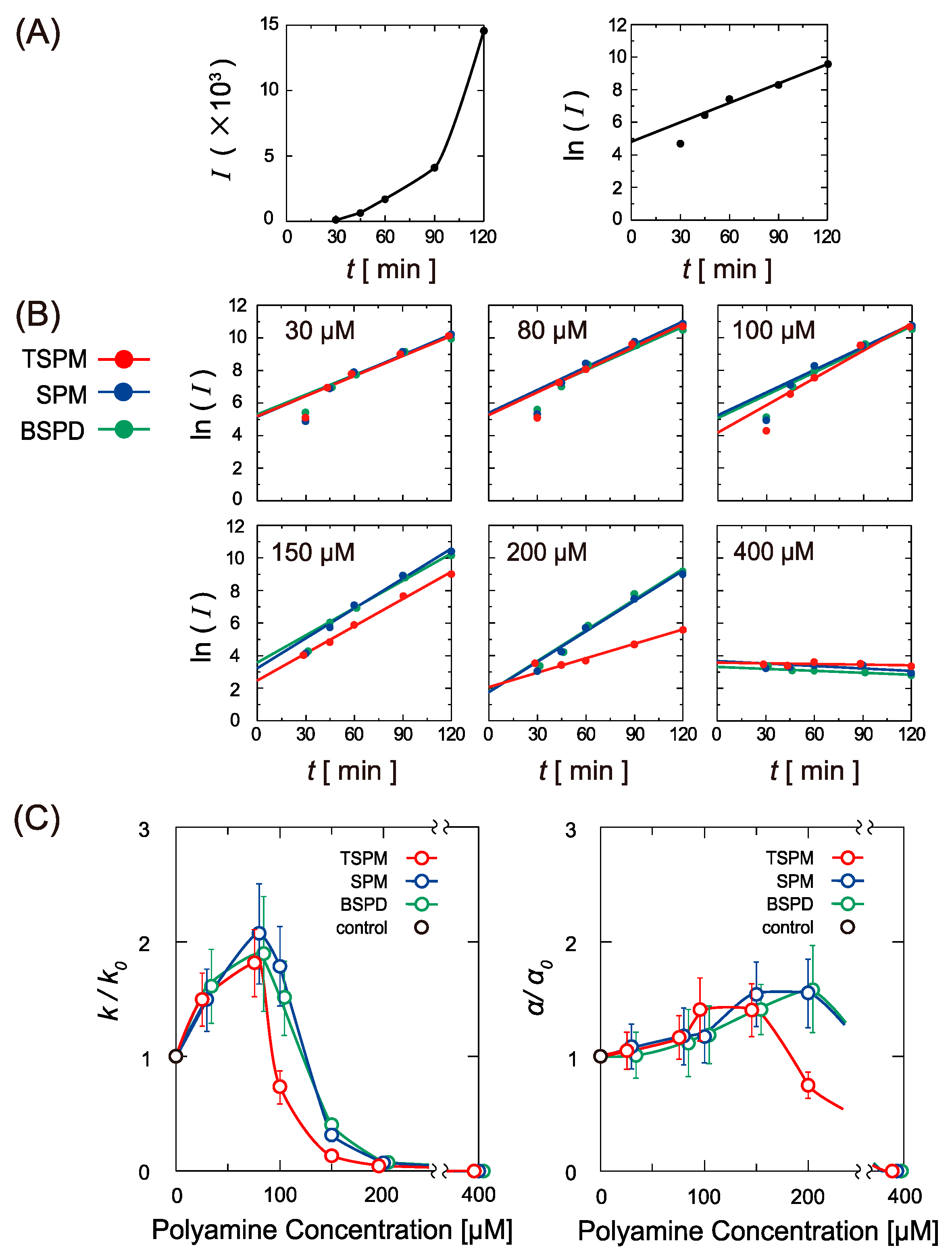
2.2. Activity of Gene Expression
2.3. Atomic Force Microscopy (AFM) Observation
2.4. Numerical Modeling of the Interaction of Polyamines with Double-Strand DNA
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Direct Observation of the Higher-Order Structure of DNA in Bulk Solution by Fluorescence Microscopy (FM)
3.2.2. Luciferase Assay for Gene Expression
3.2.3. AFM Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SPM | Spermine |
| TSPM | Thermospermine |
| BSPD | N4-Aminopropylspermidine |
| FM | Fluorescence Microscopy |
| AFM | Atomic Force Microscopy |
References
- Tabor, C.W.; Tabor, H. Polyamines in microorganisms. Microbiol. Rev. 1985, 49, 81–99. [Google Scholar] [CrossRef]
- Thomas, T.; Thomas, T. Polyamines in cell growth and cell death: Molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 2001, 58, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Childs, A.; Mehta, D.; Gerner, E. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 2003, 60, 1394–1406. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mandal, A.; Johansson, H.E.; Orjalo, A.V.; Park, M.H. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2169–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining mysteries of molecular biology: The role of polyamines in the cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 2016, 291, 14896–14903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018, 293, 18693–18701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arscott, P.G.; Li, A.Z.; Bloomfield, V.A. Condensation of DNA by trivalent cations. 1. Effects of DNA length and topology on the size and shape of condensed particles. Biopolymers 1990, 30, 619–630. [Google Scholar] [CrossRef]
- Bloomfield, V.A. Condensation of DNA by multivalent cations: Considerations on mechanism. Biopolymers 1991, 31, 1471–1481. [Google Scholar] [CrossRef]
- Bloomfield, V.A. DNA condensation by multivalent cations. Biopolymers 1997, 44, 269–282. [Google Scholar] [CrossRef]
- Vijayanathan, V.; Thomas, T.; Shirahata, A.; Thomas, T. DNA condensation by polyamines: A laser light scattering study of structural effects. Biochemistry 2001, 40, 13644–13651. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Teramoto, Y.; Yoshikawa, K. Disappearance of the Negative Charge in Giant DNA with a Folding Transition. Biophys. J. 2001, 80, 2823–2832. [Google Scholar] [CrossRef] [Green Version]
- Saminathan, M.; Thomas, T.; Shirahata, A.; Pillai, C.; Thomas, T. Polyamine structural effects on the induction and stabilization of liquid crystalline DNA: Potential applications to DNA packaging, gene therapy and polyamine therapeutics. Nucleic Acids Res. 2002, 30, 3722–3731. [Google Scholar] [CrossRef] [Green Version]
- Terui, Y.; Ohnuma, M.; Hiraga, K.; Kawashiwa, E.; Oshima, T. Stabilization of nucleic acids by unusual polyamines produced by an extreme thermophile, Thermus thermophilus. Biochem. J. 2005, 388, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igarashi, K.; Kashiwagi, K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010, 42, 39–51. [Google Scholar] [CrossRef]
- Cherstvy, A.G.; Petrov, E.P. Modeling DNA condensation on freestanding cationic lipid membranes. PCCP 2014, 16, 2020–2037. [Google Scholar] [CrossRef] [PubMed]
- Gosule, L.C.; Schellman, J.A. Compact form of DNA induced by spermidine. Nature 1976, 259, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Chattoraj, D.K.; Gosule, L.C.; Schellman, J.A. DNA condensation with polyamines: II. Electron microscopic studies. J. Mol. Biol. 1978, 121, 327–337. [Google Scholar] [CrossRef]
- Manning, G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978, 11, 179–246. [Google Scholar] [CrossRef]
- Wilson, R.W.; Bloomfield, V.A. Counterion-induced condensation of deoxyribonucleic acid. A light-scattering study. Biochemistry 1979, 18, 2192–2196. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Takahashi, M.; Vasilevskaya, V.; Khokhlov, A. Large discrete transition in a single DNA molecule appears continuous in the ensemble. Phys. Rev. Lett. 1996, 76, 3029–3031. [Google Scholar] [CrossRef] [PubMed]
- Basu, H.S.; Schwietert, H.; Feuerstein, B.; Marton, L. Effects of variation in the structure of spermine on the association with DNA and the induction of DNA conformational changes. Biochem. J. 1990, 269, 329–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, M.; Yoshikawa, K.; Vasilevskaya, V.; Khokhlov, A. Discrete coil-globule transition of single duplex DNAs induced by polyamines. J.Phys.Chem. 1997, 101, 9396–9401. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Yoshikawa, K. Diaminoalkanes with an odd number of carbon atoms induce compaction of a single double-stranded DNA chain. FEBS Lett. 1995, 361, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Zinchenko, A.A.; Sergeyev, V.G.; Yamabe, K.; Murata, S.; Yoshikawa, K. DNA compaction by divalent cations: Structural specificity revealed by the potentiality of designed quaternary diammonium salts. ChemBioChem 2004, 5, 360–368. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Umezawa, N.; Imamura, Y.; Kanbe, T.; Kato, N.; Yoshikawa, K.; Imanaka, T.; Higuchi, T. Effective chiral discrimination of tetravalent polyamines on the compaction of single DNA molecules. Angew. Chem. Int. Ed. 2013, 52, 3712–3716. [Google Scholar] [CrossRef] [PubMed]
- Pelta Jr, J.; Durand, D.; Doucet, J.; Livolant, F. DNA mesophases induced by spermidine: Structural properties and biological implications. Biophys. J. 1996, 71, 48–63. [Google Scholar] [CrossRef] [Green Version]
- Tsumoto, K.; Luckel, F.; Yoshikawa, K. Giant DNA molecules exhibit on/off switching of transcriptional activity through conformational transition. Biophys. Chem. 2003, 106, 23–29. [Google Scholar] [CrossRef]
- Luckel, F.; Kubo, K.; Tsumoto, K.; Yoshikawa, K. Enhancement and inhibition of DNA transcriptional activity by spermine: A marked difference between linear and circular templates. FEBS Lett. 2005, 579, 5119–5122. [Google Scholar] [CrossRef] [Green Version]
- Yamada, A.; Kubo, K.; Nakai, T.; Yoshikawa, K.; Tsumoto, K. All-or-none switching of transcriptional activity on single DNA molecules caused by a discrete conformational transition. Appl. Phys. Lett. 2005, 86, 223901. [Google Scholar] [CrossRef] [Green Version]
- Kanemura, A.; Yoshikawa, Y.; Fukuda, W.; Tsumoto, K.; Kenmotsu, T.; Yoshikawa, K. Opposite effect of polyamines on In vitro gene expression: Enhancement at low concentrations but inhibition at high concentrations. PLoS ONE 2018, 13, e0193595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamana, K.; Matsuzaki, S. Distinct difference in the polyamine compositions of Bryophyta and Pteridophyta. J. Biochem. 1985, 97, 1595–1601. [Google Scholar] [CrossRef]
- Bagga, S.; Rochford, J.; Klaene, Z.; Kuehn, G.D.; Phillips, G.C. Putrescine aminopropyltransferase is responsible for biosynthesis of spermidine, spermine, and multiple uncommon polyamines in osmotic stress-tolerant alfalfa. Plant. Physiol. 1997, 114, 445–454. [Google Scholar] [CrossRef] [Green Version]
- Knott, J.M.; Römer, P.; Sumper, M. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 2007, 581, 3081–3086. [Google Scholar] [CrossRef] [Green Version]
- Barra-Jimenez, A.; Ragni, L. Secondary development in the stem: When Arabidopsis and trees are closer than it seems. Curr. Opin. Plant. Biol. 2017, 35, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Kakehi, J.-i.; Kuwashiro, Y.; Niitsu, M.; Takahashi, T. Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant. Cell Physiol. 2008, 49, 1342–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, T. Unique polyamines produced by an extreme thermophile, Thermus thermophilus. Amino Acids 2007, 33, 367–372. [Google Scholar] [CrossRef]
- Wilson, Z.E.; Brimble, M.A. Molecules derives from the extremes of life. Nat. Prod. Rep. 2009, 26, 44–71. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Hidese, R.; Fukuda, W.; Niitsu, M.; Takao, K.; Horai, Y.; Umezawa, N.; Higuchi, T.; Oshima, T.; Yoshikawa, Y. Identification of a novel aminopropyltransferase involved in the synthesis of branched-chain polyamines in hyperthermophiles. J. Bacteriol. 2014, 196, 1866–1876. [Google Scholar] [CrossRef] [Green Version]
- Muramatsu, A.; Shimizu, Y.; Yoshikawa, Y.; Fukuda, W.; Umezawa, N.; Horai, Y.; Higuchi, T.; Fujiwara, S.; Imanaka, T.; Yoshikawa, K. Naturally occurring branched-chain polyamines induce a crosslinked meshwork structure in a giant DNA. J. Chem. Phys. 2016, 145, 235103. [Google Scholar] [CrossRef] [Green Version]
- Nishio, T.; Yoshikawa, Y.; Wakao Fukuda, D.; Umezawa, N.; Higuchi, T.; Fujiwara, S.; Imanaka, T.; Yoshikawa, K. Branched-Chain Polyamine Found in Hyperthermophiles Induces Unique Temperature-Dependent Structural Changes in Genome-Size DNA. ChemPhysChem 2018, 19, 2299–2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trauger, S.A.; Kallsak, E.; Kalisiak, J.; Morita, H.; Weinberg, M.V.; Menon, A.L.; Poole II, F.L.; Adam, M.W.W.; Siuzdak, G. Correlating the transcriptome, proteome, and metabolome in the environmental adaptation of hyperthermophile. J. Proteome Res. 2008, 7, 1027–1035. [Google Scholar] [CrossRef]
- Wang, M.; Yafremava, L.S.; Caerano-Anolles, D.; Mittenthal, J.E.; Caetano-Anoles, G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007, 17, 1572–1585. [Google Scholar] [CrossRef] [Green Version]
- Noireaux, V.; Bar-Ziv, R.; Libchaber, A. Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. USA 2003, 100, 12672–12677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rienzo, A.; Pascual-Ahuir, A.; Proft, M. The use of a real-time luciferase assay to quantify gene expression dynamics in the living yeast cell. Yeast 2012, 29, 219–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, M.; Faulon, J.-L.; Borkowski, O. Models for cell-free synthetic biology: Make prototyping easier, better, and faster. Front. Bioeng. Biotechnol 2018, 6, 182. [Google Scholar] [CrossRef] [PubMed]
- Nishio, T.; Yoshikawa, Y.; Shew, C.-Y.; Umezawa, N.; Higuchi, T.; Yoshikawa, K. Specific effects of antitumor active norspermidine on the structure and function of DNA. Sci. Rep. 2019, 9, 14971. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitagawa, T.; Nishio, T.; Yoshikawa, Y.; Umezawa, N.; Higuchi, T.; Shew, C.-Y.; Kenmotsu, T.; Yoshikawa, K. Effects of Structural Isomers of Spermine on the Higher-Order Structure of DNA and Gene Expression. Int. J. Mol. Sci. 2021, 22, 2355. https://doi.org/10.3390/ijms22052355
Kitagawa T, Nishio T, Yoshikawa Y, Umezawa N, Higuchi T, Shew C-Y, Kenmotsu T, Yoshikawa K. Effects of Structural Isomers of Spermine on the Higher-Order Structure of DNA and Gene Expression. International Journal of Molecular Sciences. 2021; 22(5):2355. https://doi.org/10.3390/ijms22052355
Chicago/Turabian StyleKitagawa, Tomoki, Takashi Nishio, Yuko Yoshikawa, Naoki Umezawa, Tsunehiko Higuchi, Chwen-Yang Shew, Takahiro Kenmotsu, and Kenichi Yoshikawa. 2021. "Effects of Structural Isomers of Spermine on the Higher-Order Structure of DNA and Gene Expression" International Journal of Molecular Sciences 22, no. 5: 2355. https://doi.org/10.3390/ijms22052355






