The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability
Abstract
:1. Polycomb Group Proteins and PcG Protein Complexes
2. PRC2
3. PRC1
4. Recruitment of PRCs to Target Loci
5. PcG Proteins in Human Cancer
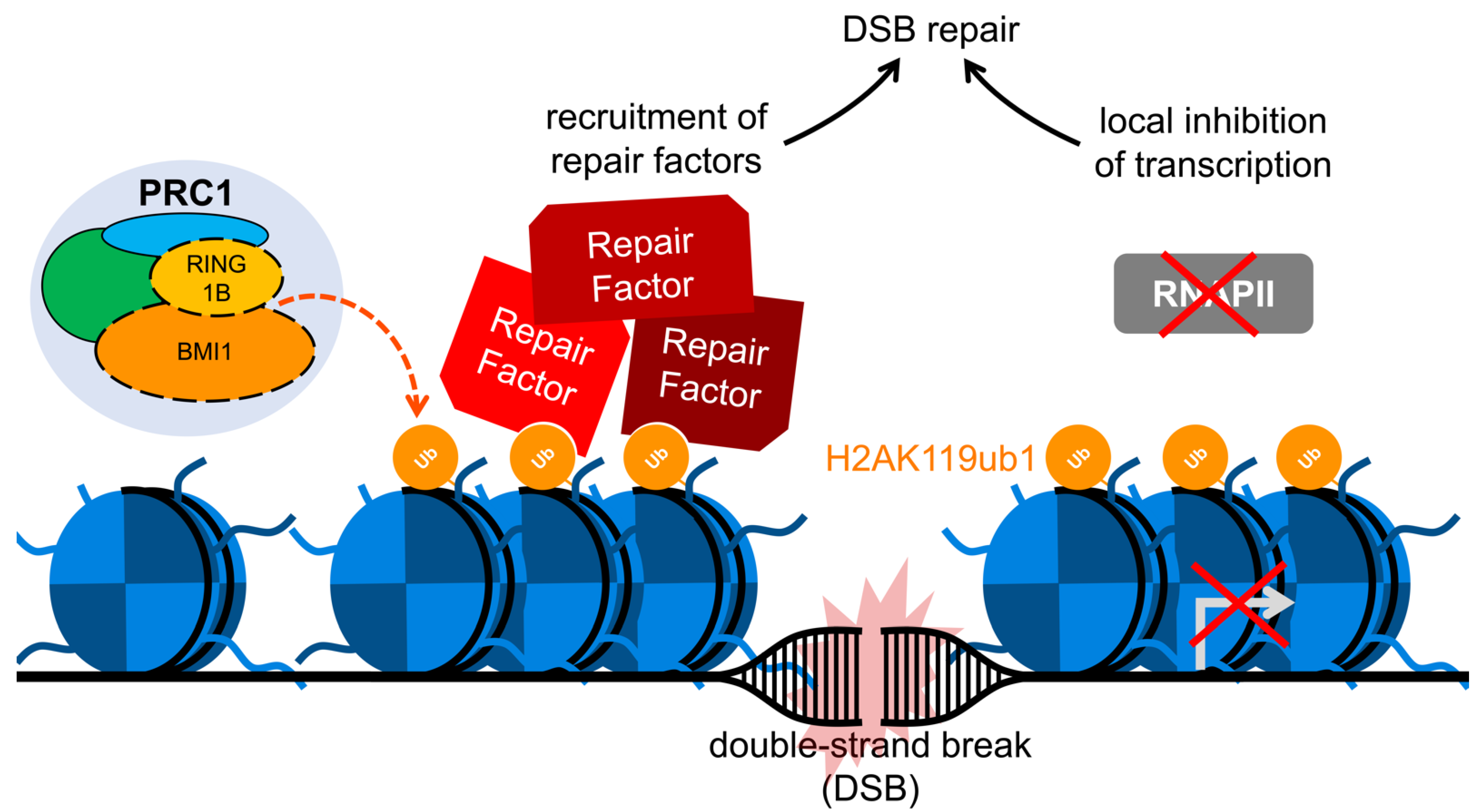
6. BMI1 and the Enhancement of Double-Strand Break Repair
7. The Link Between DNA Damage and Transcriptional Repression
8. A Role for PRC2 in Genomic Stability in Oocytes
9. Therapeutic Targeting of BMI1
Author Contributions
Funding
Conflicts of Interest
References
- Grossniklaus, U.; Paro, R. Transcriptional Silencing by Polycomb-Group Proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gearhart, M.D.; Lee, Y.W.; Kumar, I.; Ramazanov, B.; Zhang, Y.; Hernandez, C.; Lu, A.Y.; Neuenkirchen, N.; Deng, J.; et al. A Non-canonical BCOR-PRC1.1 Complex Represses Differentiation Programs in Human ESCs. Cell Stem Cell 2018, 22, 235–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duboule, D. Guidebook to the Homeobox Genes; A Sambrook and Tooze Publication at Oxford University Press: Oxford, UK, 1994; ISBN 978-0198599395. [Google Scholar]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 1981, 293, 36–41. [Google Scholar] [CrossRef]
- Tolhuis, B.; Muijrers, I.; De Wit, E.; Teunissen, H.; Talhout, W.; Van Steensel, B.; Van Lohuizen, M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006, 38, 694–699. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Kahn, T.G.; Nix, D.A.; Li, X.Y.; Bourgon, R.; Biggin, M.; Pirrotta, V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006, 38, 700–705. [Google Scholar] [CrossRef]
- Surface, L.E.; Thornton, S.R.; Boyer, L.A. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 2010, 7, 288–298. [Google Scholar] [CrossRef] [Green Version]
- Sparmann, A.; van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 2006, 6, 846–856. [Google Scholar] [CrossRef]
- Denell, R.E.; Frederick, R.D. Homoeosis in Drosophila: A description of the polycomb lethal syndrome. Dev. Biol. 1983, 97, 34–47. [Google Scholar] [CrossRef]
- Simon, J.; Chiang, A.; Bender, W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 1992, 114, 493–505. [Google Scholar]
- Pirrotta, V. Polycombing the genome: PcG, trxG and chromatin silencing. Cell 1998, 93, 333–336. [Google Scholar] [CrossRef]
- Luis, N.M.; Morey, L.; Di Croce, L.; Benitah, S.A. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell 2012, 11, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konuma, T.; Oguro, H.; Iwama, A. Role of the polycomb group proteins in hematopoietic stem cells. Dev. Growth Differ. 2010, 52, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Kassis, J.A.; Kennison, J.A.; Tamkun, J.W. Polycomb and trithorax group genes in drosophila. Genetics 2017, 206, 1699–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, T.; Verrijzer, C.P. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 2001, 20, 3055–3066. [Google Scholar] [CrossRef] [Green Version]
- Levine, S.S.; King, I.F.G.; Kingston, R.E. Division of labor in Polycomb group repression. Trends Biochem. Sci. 2004, 29, 478–485. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef]
- Cao, R.; Tsukada, Y.I.; Zhang, Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and hox gene silencing. Mol. Cell 2005, 20, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Denchi, E.L.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef] [Green Version]
- Surani, M.A.; Hayashi, K.; Hajkova, P. Genetic and epigenetic regulators of pluripotency. Cell 2007, 128, 747–762. [Google Scholar] [CrossRef] [Green Version]
- Theunissen, T.W.; Jaenisch, R. Molecular control of induced pluripotency. Cell Stem Cell 2014, 14, 720–734. [Google Scholar] [CrossRef] [Green Version]
- Ezhkova, E.; Pasolli, H.A.; Parker, J.S.; Stokes, N.; Su, I.H.; Hannon, G.; Tarakhovsky, A.; Fuchs, E. Ezh2 Orchestrates Gene Expression for the Stepwise Differentiation of Tissue-Specific Stem Cells. Cell 2009, 136, 1122–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bracken, A.P.; Kleine-Kohlbrecher, D.; Dietrich, N.; Pasini, D.; Gargiulo, G.; Beekman, C.; Theilgaard-Mönch, K.; Minucci, S.; Porse, B.T.; Marine, J.C.; et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007, 21, 525–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitcomb, S.J.; Basu, A.; Allis, C.D.; Bernstein, E. Polycomb Group proteins: An evolutionary perspective. Trends Genet. 2007, 23, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Ketel, C.S.; Andersen, E.F.; Vargas, M.L.; Suh, J.; Strome, S.; Simon, J.A. Subunit Contributions to Histone Methyltransferase Activities of Fly and Worm Polycomb Group Complexes. Mol. Cell. Biol. 2005, 25, 6857–6868. [Google Scholar] [CrossRef] [Green Version]
- Nekrasov, M.; Wild, B.; Müller, J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005, 6, 348–353. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, N.D.; Yee, D.; Chen, A.; Kalantry, S.; Chamberlain, S.J.; Otte, A.P.; Magnuson, T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005, 15, 942–947. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.A.; Kingston, R.E. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, T.L.; Zhou, B.P.; Xia, W.; Wu, Y.; Yang, C.C.; Chen-Te, C.; Ping, B.; Otte, A.P.; Hung, M.C. Molecular biology: Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005, 310, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Li, G.; Son, J.; Xu, C.F.; Margueron, R.; Neubert, T.A.; Reinberg, D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010, 24, 2615–2620. [Google Scholar] [CrossRef] [Green Version]
- Minnebo, N.; Görnemann, J.; O’Connell, N.; Van Dessel, N.; Derua, R.; Vermunt, M.W.; Page, R.; Beullens, M.; Peti, W.; Van Eynde, A.; et al. NIPP1 maintains EZH2 phosphorylation and promoter occupancy at proliferation-related target genes. Nucleic Acids Res. 2013, 41, 842–854. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.C.; Labaff, A.; Wei, Y.; Nie, L.; Xia, W.; Huo, L.; Yamaguchi, H.; Hsu, Y.H.; Hsu, J.L.; Liu, D.; et al. Phosphorylation of ezh2 at t416 by cdk2 contributes to the malignancy of triple negative breast cancers. Am. J. Transl. Res. 2015, 7, 1009–1020. [Google Scholar] [PubMed]
- Wan, L.; Xu, K.; Wei, Y.; Zhang, J.; Han, T.; Fry, C.; Zhang, Z.; Wang, Y.V.; Huang, L.; Yuan, M.; et al. Phosphorylation of EZH2 by AMPK Suppresses PRC2 Methyltransferase Activity and Oncogenic Function. Mol. Cell 2018, 69, 279–291.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Li, B.; Lin, B.; Lee, P.T.; Chung, T.H.; Tan, J.; Bi, C.; Lee, X.T.; Selvarajan, V.; Ng, S.B.; et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood 2016, 128, 948–958. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Li, G. Post-translational modifications of PRC2: Signals directing its activity. Epigenetics and Chromatin 2020, 13, 47. [Google Scholar] [CrossRef]
- Bracken, A.P.; Dietrich, N.; Pasini, D.; Hansen, K.H.; Helin, K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006, 20, 1123–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Xu, Y.; Lu, X.; Hamard, P.J.; Karl, D.L.; Man, N.; Mookhtiar, A.K.; Martinez, C.; Lossos, I.S.; Sun, J.; et al. PRMT5-mediated histone arginine methylation antagonizes transcriptional repression by polycomb complex PRC2. Nucleic Acids Res. 2020, 48, 2956–2968. [Google Scholar] [CrossRef]
- Xu, J.; Shao, Z.; Li, D.; Xie, H.; Kim, W.; Huang, J.; Taylor, J.E.; Pinello, L.; Glass, K.; Jaffe, J.D.; et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 2015, 57, 304–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.H.; Kim, W.; Howard, T.P.; Vazquez, F.; Tsherniak, A.; Wu, J.N.; Wang, W.; Haswell, J.R.; Walensky, L.D.; Hahn, W.C.; et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 2015, 21, 1491–1496. [Google Scholar] [CrossRef] [Green Version]
- Svrlanska, A.; Reichel, A.; Schilling, E.-M.; Scherer, M.; Stamminger, T.; Reuter, N. A Noncanonical Function of Polycomb Repressive Complexes Promotes Human Cytomegalovirus Lytic DNA Replication and Serves as a Novel Cellular Target for Antiviral Intervention. J. Virol. 2019, 93, e02143-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, C.L.; Baldwin, A.S. Non-canonical EZH2 transcriptionally activates RelB in triple negative breast cancer. PLoS ONE 2016, 11, e0165005. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.T.; Li, Z.; Wu, Z.; Aau, M.; Guan, P.; Karuturi, R.K.M.; Liou, Y.C.; Yu, Q. Context-Specific Regulation of NF-κB Target Gene Expression by EZH2 in Breast Cancers. Mol. Cell 2011, 43, 798–810. [Google Scholar] [CrossRef]
- Mousavi, K.; Zare, H.; Wang, A.H.; Sartorelli, V. Polycomb Protein Ezh1 Promotes RNA Polymerase II Elongation. Mol. Cell 2012, 45, 255–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healy, E.; Mucha, M.; Glancy, E.; Fitzpatrick, D.J.; Conway, E.; Neikes, H.K.; Monger, C.; Van Mierlo, G.; Baltissen, M.P.; Koseki, Y.; et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cell 2019, 76, 437–452. [Google Scholar] [CrossRef]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856. [Google Scholar] [CrossRef] [Green Version]
- van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019, 29, 660–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alekseyenko, A.A.; Gorchakov, A.A.; Kharchenko, P.V.; Kuroda, M.I. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. USA 2014, 111, 2488–2493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smits, A.H.; Jansen, P.W.T.C.; Poser, I.; Hyman, A.A.; Vermeulen, M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013, 41, e28. [Google Scholar] [CrossRef]
- van Mierlo, G.; Dirks, R.A.M.; De Clerck, L.; Brinkman, A.B.; Huth, M.; Kloet, S.L.; Saksouk, N.; Kroeze, L.I.; Willems, S.; Farlik, M.; et al. Integrative Proteomic Profiling Reveals PRC2-Dependent Epigenetic Crosstalk Maintains Ground-State Pluripotency. Cell Stem Cell 2019, 24, 123–137. [Google Scholar] [CrossRef] [Green Version]
- Holoch, D.; Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017, 42, 531–542. [Google Scholar] [CrossRef]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun 2016, 7, 13661. [Google Scholar] [CrossRef] [Green Version]
- Gil, J.; Peters, G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol. 2006, 7, 667–677. [Google Scholar] [CrossRef]
- Buchwald, G.; Van Der Stoop, P.; Weichenrieder, O.; Perrakis, A.; Van Lohuizen, M.; Sixma, T.K. Structure and E3-ligase activity of the Ring-Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 2006, 25, 2465–2474. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahn, T.G.; Dorafshan, E.; Schultheis, D.; Zare, A.; Stenberg, P.; Reim, I.; Pirrotta, V.; Schwartz, Y.B. Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res. 2016, 44, 10132–10149. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [Green Version]
- Morey, L.; Santanach, A.; Blanco, E.; Aloia, L.; Nora, E.P.; Bruneau, B.G.; Di croce, L. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell 2015, 17, 300–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loubiere, V.; Delest, A.; Thomas, A.; Bonev, B.; Schuettengruber, B.; Sati, S.; Martinez, A.M.; Cavalli, G. Coordinate redeployment of PRC1 proteins suppresses tumor formation during Drosophila development. Nat. Genet. 2016, 48, 1436–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between polycomb repressive complexes. Elife 2016, 5, e18591. [Google Scholar] [CrossRef]
- Grau, D.J.; Chapman, B.A.; Garlick, J.D.; Borowsky, M.; Francis, N.J.; Kingston, R.E. Compaction of chromatin by diverse polycomb group proteins requires localized regions of high charge. Genes Dev. 2011, 25, 2210–2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neira, J.L.; Román-Trufero, M.; Contreras, L.M.; Prieto, J.; Singh, G.; Barrera, F.N.; Renart, M.L.; Vidal, M. The transcriptional repressor RYBP is a natively unfolded protein which folds upon binding to DNA. Biochemistry 2009, 48, 1348–1360. [Google Scholar] [CrossRef]
- Freemont, P.S.; Hanson, I.M.; Trowsdale, J. A novel gysteine-rich sequence motif. Cell 1991, 64, 483–484. [Google Scholar] [CrossRef]
- Vidal, M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int. J. Dev. Biol. 2009, 53, 355–370. [Google Scholar] [CrossRef] [Green Version]
- Czypionka, A.; De Los Paños, O.R.; Mateu, M.G.; Barrera, F.N.; Hurtado-Gómez, E.; Gómez, J.; Vidal, M.; Neira, J.L. The isolated C-terminal domain of ring 1B is a dimer made of stable, well-structured monomers. Biochemistry 2007, 46, 12764–12776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb group proteins ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Alchanati, I.; Teicher, C.; Cohen, G.; Shemesh, V.; Barr, H.M.; Nakache, P.; Ben-Avraham, D.; Idelevich, A.; Angel, I.; Livnah, N.; et al. The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2α cleavage complex—A potentially new drug target. PLoS ONE 2009, 4, e8104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Mar Lorente, M.; Marcos-Gutierrez, C.; Perez, C.; Schoorlemmer, J.; Ramirez, A.; Magin, T.; Vidal, M. Loss- and gain-of-function mutations show a Polycomb group function for Ring1A in mice. Development 2000, 127, 5093–5100. [Google Scholar]
- Illingworth, R.S.; Moffat, M.; Mann, A.R.; Read, D.; Hunter, C.J.; Pradeepa, M.M.; Adams, I.R.; Bickmore, W.A. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015, 29, 1897–1902. [Google Scholar] [CrossRef] [Green Version]
- Leeb, M.; Wutz, A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 2007, 178, 219–229. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874. [Google Scholar] [CrossRef] [Green Version]
- Scheuermann, J.C.; De Ayala Alonso, A.G.; Oktaba, K.; Ly-Hartig, N.; McGinty, R.K.; Fraterman, S.; Wilm, M.; Muir, T.W.; Müller, J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010, 465, 243–247. [Google Scholar] [CrossRef] [Green Version]
- Chittock, E.C.; Latwiel, S.; Miller, T.C.R.; Müller, C.W. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017, 45, 193–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellino, G.I.; Schwartz, Y.B.; Farkas, G.; McCabe, D.; Elgin, S.C.R.; Pirrotta, V. Polycomb silencing blocks transcription initiation. Mol. Cell 2004, 13, 887–893. [Google Scholar] [CrossRef] [Green Version]
- King, I.F.G.; Francis, N.J.; Kingston, R.E. Native and Recombinant Polycomb Group Complexes Establish a Selective Block to Template Accessibility To Repress Transcription In Vitro. Mol. Cell. Biol. 2002, 22, 7919–7928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morey, L.; Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Pherson, M.; Misulovin, Z.; Gause, M.; Mihindukulasuriya, K.; Swain, A.; Dorsett, D. Polycomb repressive complex 1 modifies transcription of active genes. Sci. Adv. 2017, 3, e1700944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaaf, C.A.; Misulovin, Z.; Gause, M.; Koenig, A.; Gohara, D.W.; Watson, A.; Dorsett, D. Cohesin and Polycomb Proteins Functionally Interact to Control Transcription at Silenced and Active Genes. PLoS Genet. 2013, 9, e1003560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandamme, J.; Völkel, P.; Rosnoblet, C.; Le Faou, P.; Angrand, P.O. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol. Cell. Proteomics 2011, 10, M110-002642. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, H.; Ishiguro, K.I.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A complex with chromatin modifiers that occupies E2f- and Myc-responsive genes in G0 cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef]
- Kerppola, T.K. Polycomb group complexes--many combinations, many functions. Trends Cell Biol. 2009, 19, 692–704. [Google Scholar] [CrossRef] [Green Version]
- Müller, J.; Kassis, J.A. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 2006, 16, 476–484. [Google Scholar] [CrossRef]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Jenner, R.G.; Boyer, L.A.; Guenther, M.G.; Levine, S.S.; Kumar, R.M.; Chevalier, B.; Johnstone, S.E.; Cole, M.F.; Isono, K.I.; et al. Control of Developmental Regulators by Polycomb in Human Embryonic Stem Cells. Cell 2006, 125, 301–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, M.; Koche, R.P.; Rheinbay, E.; Mendenhall, E.M.; Endoh, M.; Mikkelsen, T.S.; Presser, A.; Nusbaum, C.; Xie, X.; Chi, A.S.; et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008, 4, e1000242. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Cavalli, G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009, 136, 3531–3542. [Google Scholar] [CrossRef] [Green Version]
- Caretti, G.; Di Padova, M.; Micales, B.; Lyons, G.E.; Sartorelli, V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004, 18, 2627–2638. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef]
- Portoso, M.; Ragazzini, R.; Brenčič, Ž.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017, 36, 981–994. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [Green Version]
- Obier, N.; Cauchy, P. Polycomb at the crossroads: An integrated model with lncRNA- And silencer-mediated repression. J. RNA Genomics 2018, 14, 592–605. [Google Scholar]
- Lavarone, E.; Barbieri, C.M.; Pasini, D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat. Commun. 2019, 10, 1679. [Google Scholar] [CrossRef]
- Garcia, E.; Marcos-Gutiérrez, C.; Del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.M.; Demeler, B.; et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 2010, 18, 966–975. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Wang, M.; Chang, L.; Yu, J.; Song, A.; Liu, C.; Huang, W.; Zhang, T.; Wu, X.; Shen, X.; et al. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat. Cell Biol. 2020, 22, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Trojer, P.; Cao, A.R.; Gao, Z.; Li, Y.; Zhang, J.; Xu, X.; Li, G.; Losson, R.; Erdjument-Bromage, H.; Tempst, P.; et al. L3MBTL2 Protein Acts in Concert with PcG Protein-Mediated Monoubiquitination of H2A to Establish a Repressive Chromatin Structure. Mol. Cell 2011, 42, 438–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalenik, J.L.; Chen, D.; Bradley, M.E.; Chen, S.J.; Lee, T.C. Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 1997, 25, 843–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, F.; Pratt, H.; Atchison, M.L. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J. Cell. Biochem. 2010, 109, 478–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendenhall, E.M.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010, 6, e1001244. [Google Scholar] [CrossRef] [Green Version]
- Zeisig, B.B.; So, C.W.E. Combinatorial Tethering: A Novel Mode to Recruit Non-canonical PRC1 for Normal and Malignant GC B Cell Development. Cancer Cell 2016, 30, 185–187. [Google Scholar] [CrossRef] [Green Version]
- Haupt, Y.; Alexander, W.S.; Barri, G.; Peter Klinken, S.; Adams, J.M. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in Eμ-myc transgenic mice. Cell 1991, 65, 753–763. [Google Scholar] [CrossRef]
- Van Lohuizen, M.; Frasch, M.; Wientjens, E.; Berns, A. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 1991, 353, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.J.L.; Scheijen, B.; Voncken, J.W.; Kieboom, K.; Berns, A.; Van Lohuizen, M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc- induced apoptosis via INK4a/ARF. Genes Dev. 1999, 13, 2678–2690. [Google Scholar] [CrossRef]
- Sharpless, N.E.; Depinho, R.A. The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 1999, 9, 22–30. [Google Scholar] [CrossRef]
- Ouelle, D.E.; Zindy, F.; Ashmun, R.A.; Sherr, C.J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 1995, 83, 993–1000. [Google Scholar] [CrossRef] [Green Version]
- Hudson, C.D.; Morris, P.J.; Latchman, D.S.; Budhram-Mahadeo, V.S. Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Bax in vitro and in vivo to determine cell fate. J. Biol. Chem. 2005, 280, 11851–11858. [Google Scholar] [CrossRef] [Green Version]
- Dovey, J.S.; Zacharek, S.J.; Kim, C.F.; Lees, J.A. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc. Natl. Acad. Sci. USA 2008, 105, 11857–11862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itahana, K.; Zou, Y.; Itahana, Y.; Martinez, J.-L.; Beausejour, C.; Jacobs, J.J.L.; van Lohuizen, M.; Band, V.; Campisi, J.; Dimri, G.P. Control of the Replicative Life Span of Human Fibroblasts by p16 and the Polycomb Protein Bmi-1. Mol. Cell. Biol. 2003, 23, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Dimri, G.P.; Martinez, J.L.; Jacobs, J.J.L.; Keblusek, P.; Itahana, K.; Van Lohuizen, M.; Campisi, J.; Wazer, D.E.; Band, V. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 2002, 62, 4736–4745. [Google Scholar]
- Kim, J.H.; Yoon, S.Y.; Kim, C.N.; Joo, J.H.; Moon, S.K.; Choe, I.S.; Choe, Y.K.; Kim, J.W. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004, 203, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Lingbeek, M.; Shakhova, O.; Liu, J.; Tanger, E.; Saremaslani, P.; Van Lohuizen, M.; Marino, S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 2004. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, L.; Wan, G.; Dou, T.; Tian, J. BMI1 promotes the progression of laryngeal squamous cell carcinoma. Oral Oncol. 2011, 47, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yoon, S.Y.; Jeong, S.H.; Kim, S.Y.; Moon, S.K.; Joo, J.H.; Lee, Y.; Choe, I.S.; Kim, J.W. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast 2004, 13, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, R.U.; Memarzadeh, S.; Wu, H.; Witte, O.N. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell 2010, 7, 682–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, E.; Waghray, M.; Lee, C.J.; Heidt, D.G.; Yalamanchili, M.; Li, C.; Bednar, F.; Simeone, D.M. Bmi1 Enhances Tumorigenicity and Cancer Stem Cell Function in Pancreatic Adenocarcinoma. PLoS ONE 2013, 8, e55820. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; He, L.; Kapoor, A.; Rybak, A.P.; De Melo, J.; Cutz, J.C.; Tang, D. PTEN inhibits BMI1 function independently of its phosphatase activity. Mol. Cancer 2009, 8, 98. [Google Scholar] [CrossRef] [Green Version]
- Haupt, Y.; Bath, M.L.; Harris, A.W.; Adams, J.M. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene 1993, 8, 3161–3164. [Google Scholar]
- Alkema, M.J.; Jacobs, H.; Van Lohuizen, M.; Berns, A. Perturbation of B and T cell development and predisposition to lymphomagenesis in EμBmi1 transgenic mice require the Bmi1 RING finger. Oncogene 1997, 15, 899–910. [Google Scholar] [CrossRef] [Green Version]
- Lessard, J.; Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003, 423, 255–260. [Google Scholar] [CrossRef]
- Beà, S.; Tort, F.; Pinyol, M.; Puig, X.; Hernández, L.; Hernández, S.; Fernández, P.L.; Van Lohuizen, M.; Colomer, D.; Campo, E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001, 61, 2409–2412. [Google Scholar]
- Vonlanthen, S.; Heighway, J.; Altermatt, H.J.; Gugger, M.; Kappeler, A.; Borner, M.M.; van Lohuizen, M.; Betticher, D.C. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br. J. Cancer 2001, 84, 1372–1376. [Google Scholar] [CrossRef] [Green Version]
- Koren, A.; Rijavec, M.; Sodja, E.; Kern, I.; Sadikov, A.; Kovac, V.; Korosec, P.; Cufer, T. High BMI1 mRNA expression in peripheral whole blood is associated with favorable prognosis in advanced non-small cell lung cancer patients. Oncotarget 2017, 8, 25384–25394. [Google Scholar] [CrossRef] [Green Version]
- Berezovska, O.P.; Glinskii, A.B.; Yang, Z.; Li, X.M.; Hoffman, R.M.; Glinsky, G.V. Essential role for activation of the polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle 2006, 5, 1886–1901. [Google Scholar] [CrossRef] [Green Version]
- van Leenders, G.J.L.H.; Dukers, D.; Hessels, D.; van den Kieboom, S.W.M.; Hulsbergen, C.A.; Witjes, J.A.; Otte, A.P.; Meijer, C.J.; Raaphorst, F.M. Polycomb-Group Oncogenes EZH2, BMI1, and RING1 Are Overexpressed in Prostate Cancer With Adverse Pathologic and Clinical Features. Eur. Urol. 2007, 52, 455–463. [Google Scholar] [CrossRef]
- Fan, C.; He, L.; Kapoor, A.; Gillis, A.; Rybak, A.P.; Cutz, J.C.; Tang, D. Bmi1 promotes prostate tumorigenesis via inhibiting p16INK4A and p14ARF expression. Biochim. Biophys. Acta-Mol. Basis Dis. 2008, 1782, 642–648. [Google Scholar] [CrossRef] [Green Version]
- Glinsky, G.V.; Berezovska, O.; Glinskii, A.B. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 2005, 115, 1503–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, L.; Bombard, J.; Cintron, K.; Sheedy, J.; Weetall, M.L.; Davis, T.W. BMI1 as a novel target for drug discovery in cancer. J. Cell. Biochem. 2011, 112, 2729–2741. [Google Scholar] [CrossRef]
- Abdouh, M.; Facchino, S.; Chatoo, W.; Balasingam, V.; Ferreira, J.; Bernier, G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J. Neurosci. 2009, 29, 8884–8896. [Google Scholar] [CrossRef]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; RayChaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by MicroRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef] [Green Version]
- Korur, S.; Huber, R.M.; Sivasankaran, B.; Petrich, M.; Morin, P.; Hemmings, B.A.; Merlo, A.; Lino, M.M. GSK3β Regulates Differentiation and Growth Arrest in Glioblastoma. PLoS ONE 2009, 4, e7443. [Google Scholar] [CrossRef] [PubMed]
- Suvà, M.L.; Riggi, N.; Janiszewska, M.; Radovanovic, I.; Provero, P.; Stehle, J.C.; Baumer, K.; Le Bitoux, M.A.; Marino, D.; Cironi, L.; et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009, 69, 9211–9218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breuer, R.H.J.; Snijders, P.J.F.; Smit, E.F.; Sutedja, T.G.; Sewalt, R.G.A.B.; Otte, A.P.; Van Kemenade, F.J.; Postmus, P.E.; Meijer, C.J.L.M.; Raaphorst, F.M. Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia 2004, 6, 736–743. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.K.; Kim, R.H.; Kim, S.J.; Yip, F.K.; Shin, K.H.; Dimri, G.P.; Christensen, R.; Han, T.; Park, N.H. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br. J. Cancer 2007, 96, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Ikeda, H.; Itatsu, K.; Yamaguchi, J.; Sawada, S.; Minato, H.; Ohta, T.; Nakanuma, Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab. Investig. 2008, 88, 873–882. [Google Scholar] [CrossRef]
- Steele, J.C.; Torr, E.E.; Noakes, K.L.; Kalk, E.; Moss, P.A.; Reynolds, G.M.; Hubscher, S.G.; Van Lohuizen, M.; Adams, D.H.; Young, L.S. The polycomb group proteins, BMI-1 and EZH2, are tumour-associated antigens. Br. J. Cancer 2006, 95, 1202–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Zhang, N.; Zheng, Y.; De Wilde, R.F.; Maitra, A.; Pan, D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010, 24, 2383–2388. [Google Scholar] [CrossRef] [Green Version]
- Mimori, K.; Sadanaga, N.; Yoshikawa, Y.; Ishikawa, K.; Hashimoto, M.; Tanaka, F.; Sasaki, A.; Inoue, H.; Sugimachi, K.; Mori, M. Reduced tau expression in gastric cancer can identify candidates for successful Paclitaxel treatment. Br. J. Cancer 2006, 94, 1894–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tateishi, K.; Ohta, M.; Kanai, F.; Guleng, B.; Tanaka, Y.; Asaoka, Y.; Tada, M.; Seto, M.; Jazag, A.; Lianjie, L.; et al. Dysregulated expression of stem cell factor Bmi1 in precancerous lesions of the gastrointestinal tract. Clin. Cancer Res. 2006, 12, 6960–6966. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, H.; Iyer, S.V.; Sasaki, K.; Tawfik, O.W.; Iwakuma, T. An improved intrafemoral injection with minimized leakage as an –orthotopic mouse model of osteosarcoma. Anal. Biochem. 2015, 486, 70–74. [Google Scholar] [CrossRef] [Green Version]
- Arisan, S.; Buyuktuncer, E.D.; Palavan-Unsal, N.; Çaşkurlu, T.; Çakir, O.O.; Ergenekon, E. Increased expression of EZH2, a polycomb group protein, in bladder carcinoma. Urol. Int. 2005, 75, 252–257. [Google Scholar] [CrossRef]
- Raman, J.D.; Mongan, N.P.; Tickoo, S.K.; Boorjian, S.A.; Scherr, D.S.; Gudas, L.J. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin. Cancer Res. 2005, 11, 8570–8576. [Google Scholar] [CrossRef] [Green Version]
- Shafaroudi, A.M.; Mowla, S.J.; Ziaee, S.A.M.; Bahrami, A.R.; Atlasi, Y.; Malakootian, M. Overexpression of BMI1, a polycomb group repressor protein, in bladder tumors: A preliminary report. Urol. J. 2008, 13, 277. [Google Scholar] [CrossRef]
- Weikert, S.; Christoph, F.; Köllermann, J.; Müller, M.; Schrader, M.; Miller, K.; Krause, H. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int. J. Mol. Med. 2005, 16, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.A.B.; Rubin, M.A.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Bracken, A.P.; Pasini, D.; Capra, M.; Prosperini, E.; Colli, E.; Helin, K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003, 22, 5323–5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, D.; Martinez-Leal, J.F.; Rizzo, S.; Martinez, D.; Hudson, D.; Visakorpi, T.; Peters, G.; Carnero, A.; Beach, D.; Gil, J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene 2005, 24, 5543–5551. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Cao, Q.; Mehra, R.; Laxman, B.; Yu, J.; Tomlins, S.A.; Creighton, C.J.; Dhanasekaran, S.M.; Shen, R.; Chen, G.; et al. Integrative Genomics Analysis Reveals Silencing of β-Adrenergic Signaling by Polycomb in Prostate Cancer. Cancer Cell 2007, 12, 419–431. [Google Scholar] [CrossRef] [Green Version]
- Collett, K.; Eide, G.E.; Arnes, J.; Stefansson, I.M.; Eide, J.; Braaten, A.; Aas, T.; Otte, A.P.; Akslen, L.A. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin. Cancer Res. 2006, 12, 1168–1174. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Erdmann, C.; Chinnaiyan, A.M.; Merajver, S.D.; Kleer, C.G. Identification of EZH2 as a molecular marker for a precancerous state in morphologically normal breast tissues. Cancer Res. 2006, 66, 4095–4099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puppe, J.; Drost, R.; Liu, X.; Joosse, S.A.; Evers, B.; Cornelissen-Steijger, P.; Nederlof, P.; Yu, Q.; Jonkers, J.; van Lohuizen, M.; et al. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb Repressive Complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009, 11, R63. [Google Scholar] [CrossRef] [Green Version]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.A.B.; Otte, A.P.; Hayes, D.F.; et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joensuu, K.; Hagström, J.; Leidenius, M.; Haglund, C.; Andersson, L.C.; Sariola, H.; Heikkilä, P. Bmi-1, c-myc, and Snail expression in primary breast cancers and their metastases-elevated Bmi-1 expression in late breast cancer relapses. Virchows Arch. 2011, 459, 31–39. [Google Scholar] [CrossRef]
- Hoenerhoff, M.J.; Chu, I.; Barkan, D.; Liu, Z.Y.; Datta, S.; Dimri, G.P.; Green, J.E. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene 2009, 28, 3022–3032. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.F.; Sun, Q.Y.; Ding, L.W.; Chien, W.; Liu, X.Y.; Mayakonda, A.; Jiang, Y.Y.; Loh, X.Y.; Ran, X.B.; Doan, N.B.; et al. The c-MYC–BMI1 axis is essential for SETDB1-mediated breast tumourigenesis. J. Pathol. 2018, 246, 89–102. [Google Scholar] [CrossRef]
- Esteller, M. CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene 2002, 21, 5427–5440. [Google Scholar] [CrossRef] [Green Version]
- Curtin, K.; Slattery, M.L.; Samowitz, W.S. CpG Island Methylation in Colorectal Cancer: Past, Present and Future. Patholog. Res. Int. 2011, 2011, 902674. [Google Scholar] [CrossRef] [Green Version]
- Herman, J.G.; Latif, F.; Weng, Y.; Lerman, M.I.; Zbar, B.; Liu, S.; Samid, D.; Duan, D.S.R.; Gnarra, J.R.; Linehan, W.M.; et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. USA 1994, 91, 9700–9704. [Google Scholar] [CrossRef] [Green Version]
- Kaise, M.; Yamasaki, T.; Yonezawa, J.; Miwa, J.; Ohta, Y.; Tajiri, H. CpG island hypermethylation of tumor-suppressor genes in H. pylori-infected non-neoplastic gastric mucosa is linked with gastric cancer risk. Helicobacter 2008, 13, 35–41. [Google Scholar] [CrossRef]
- Veneti, Z.; Gkouskou, K.K.; Eliopoulos, A.G. Polycomb repressor complex 2 in genomic instability and cancer. Int. J. Mol. Sci. 2017, 18, 1657. [Google Scholar] [CrossRef] [Green Version]
- Casanova, M.; Preissner, T.; Cerase, A.; Poot, R.; Yamada, D.; Li, X.; Appanah, R.; Bezstarosti, K.; Demmers, J.; Koseki, H.; et al. Polycomblike 2 facilitates the recruitment of PRC2 Polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development 2011, 138, 1471–1482. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liefke, R.; Jiang, J.; Kurland, J.V.; Tian, W.; Deng, P.; Zhang, W.; He, Q.; Patel, D.J.; Bulyk, M.L.; et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 2017, 549, 287–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perino, M.; Van Mierlo, G.; Karemaker, I.D.; Van Genesen, S.; Vermeulen, M.; Marks, H.; Van Heeringen, S.J.; Veenstra, G.J.C. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018, 50, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Bergink, S.; Salomons, F.A.; Hoogstraten, D.; Groothuis, T.A.M.; De Waard, H.; Wu, J.; Yuan, L.; Citterio, E.; Houtsmuller, A.B.; Neefjes, J.; et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006, 20, 1343–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair 2017, 56, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.H.; Andrin, C.; McDonald, D.; Hendzel, M.J. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol 2010, 191, 45–60. [Google Scholar] [CrossRef]
- Pan, M.R.; Peng, G.; Hung, W.C.; Lin, S.Y.; Hung, W.C.; Lin, S.Y. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol Chem 2011, 286, 28599–28607. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Cao, R.; Wang, M.; Myers, M.P.; Zhang, Y.; Xu, R.M. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 2006, 281, 20643–20649. [Google Scholar] [CrossRef] [Green Version]
- Facchino, S.; Abdouh, M.; Chatoo, W.; Bernier, G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 2010, 30, 10096–10111. [Google Scholar] [CrossRef]
- Chagraoui, J.; Hébert, J.; Girard, S.; Sauvageau, G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc. Natl. Acad. Sci. USA 2011, 108, 5284–5289. [Google Scholar] [CrossRef] [Green Version]
- Ginjala, V.; Nacerddine, K.; Kulkarni, A.; Oza, J.; Hill, S.J.; Yao, M.; Citterio, E.; van Lohuizen, M.; Ganesan, S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell Biol 2011, 31, 1972–1982. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.Y.; Kang, H.Y.; Yang, W.L.; Wu, J.; Jeong, Y.S.; Wang, J.; Chan, C.H.; Lee, S.W.; Zhang, X.; Lamothe, B.; et al. Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J. Biol Chem 2011, 286, 30806–30815. [Google Scholar] [CrossRef] [Green Version]
- Chou, D.M.; Adamson, B.; Dephoure, N.E.; Tan, X.; Nottke, A.C.; Hurov, K.E.; Gygi, S.P.; Colaiácovo, M.P.; Elledge, S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 2010, 107, 18475–18480. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.R.; Sossick, A.J.; Boulton, S.J.; Jackson, S.P. BRCA1-associated exclusion of 53BP1 from DNA: Damage sites underlies temporal control of DNA repair. J. Cell Sci. 2012, 125, 3529–3534. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.M. BRCA1 in the DNA damage response and at telomeres. Front. Genet. 2013, 4, 85. [Google Scholar] [CrossRef] [Green Version]
- Daley, J.M.; Sung, P. 53BP1, BRCA1, and the Choice between Recombination and End Joining at DNA Double-Strand Breaks. Mol. Cell. Biol. 2014, 34, 1380–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartocci, C.; Diedrich, J.K.; Ouzounov, I.; Li, J.; Piunti, A.; Pasini, D.; Yates, J.R.; Lazzerini Denchi, E. Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep. 2014, 7, 1320–1332. [Google Scholar] [CrossRef] [Green Version]
- Gieni, R.S.; Ismail, I.H.; Campbell, S.; Hendzel, M.J. Polycomb group proteins in the DNA damage response: A link between radiation resistance and “stemness. ” Cell Cycle 2011, 10, 883–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemenway, C.S.; Halligan, B.W.; Levy, L.S. The Bmi-1 oncoprotein interacts with dinG and MPh2: The role of RING finger domains. Oncogene 1998, 16, 2541–2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattiroli, F.; Vissers, J.H.A.; Van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef] [Green Version]
- Leung, J.W.; Agarwal, P.; Canny, M.D.; Gong, F.; Robison, A.D.; Finkelstein, I.J.; Durocher, D.; Miller, K.M. Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling. PLoS Genet. 2014, 10, e1004178. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-T.; Alpert, A.; Leiter, C.; Gong, F.; Jackson, S.P.; Miller, K.M. Systematic Identification of Functional Residues in Mammalian Histone H2AX. Mol. Cell. Biol. 2013, 33, 111–126. [Google Scholar] [CrossRef] [Green Version]
- McGinty, R.K.; Henrici, R.C.; Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 2014, 514, 591–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakarougkas, A.; Ismail, A.; Chambers, A.L.; Riballo, E.; Herbert, A.D.; Kunzel, J.; Lobrich, M.; Jeggo, P.A.; Downs, J.A. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 2014, 55, 723–732. [Google Scholar] [CrossRef] [Green Version]
- Nacerddine, K.; Beaudry, J.B.; Ginjala, V.; Westerman, B.; Mattiroli, F.; Song, J.Y.; Van Der Poel, H.; Ponz, O.B.; Pritchard, C.; Cornelissen-Steijger, P.; et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J. Clin. Invest. 2012, 122, 1920–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puc, J.; Kozbial, P.; Li, W.; Tan, Y.; Liu, Z.; Suter, T.; Ohgi, K.A.; Zhang, J.; Aggarwal, A.K.; Rosenfeld, M.G. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 2015, 160, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Periyasamy, M.; Patel, H.; Lai, C.F.; Nguyen, V.T.M.; Nevedomskaya, E.; Harrod, A.; Russell, R.; Remenyi, J.; Ochocka, A.M.; Thomas, R.S.; et al. APOBEC3B-Mediated Cytidine Deamination Is Required for Estrogen Receptor Action in Breast Cancer. Cell Rep. 2015, 13, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Vitelli, V.; Galbiati, A.; Iannelli, F.; Pessina, F.; Sharma, S.; D’Adda di Fagagna, F. Recent Advancements in DNA Damage-Transcription Crosstalk and High-Resolution Mapping of DNA Breaks. Annu. Rev. Genomics Hum. Genet. 2017, 18, 87–113. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, J.M.R.; Yang, Y.G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Shanbhag, N.M.; Rafalska-Metcalf, I.U.; Balane-Bolivar, C.; Janicki, S.M.; Greenberg, R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010, 141, 970–981. [Google Scholar] [CrossRef] [Green Version]
- Iannelli, F.; Galbiati, A.; Capozzo, I.; Nguyen, Q.; Magnuson, B.; Michelini, F.; D’Alessandro, G.; Cabrini, M.; Roncador, M.; Francia, S.; et al. A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks. Nat. Commun. 2017, 8, 15656. [Google Scholar] [CrossRef]
- Kim, J.; Sturgill, D.; Tran, A.D.; Sinclair, D.A.; Oberdoerffer, P. Controlled DNA double-strand break induction in mice reveals post-damage transcriptome stability. Nucleic Acids Res. 2015, 44, e64. [Google Scholar] [CrossRef] [Green Version]
- Adam, S.; Polo, S.E. Blurring the line between the DNA damage response and transcription: The importance of chromatin dynamics. Exp. Cell Res. 2014, 329, 148–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankotai, T.; Bonhomme, C.; Chen, D.; Soutoglou, E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012, 19, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Harding, S.M.; Boiarsky, J.A.; Greenberg, R.A. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep. 2015, 13, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; De Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; D’adda Di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Becherel, O.J.; Yeo, A.J.; Stellati, A.; Heng, E.Y.H.; Luff, J.; Suraweera, A.M.; Woods, R.; Fleming, J.; Carrie, D.; McKinney, K.; et al. Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing. PLoS Genet. 2013, 9, e1003435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beli, P.; Lukashchuk, N.; Wagner, S.A.; Weinert, B.T.; Olsen, J.V.; Baskcomb, L.; Mann, M.; Jackson, S.P.; Choudhary, C. Proteomic Investigations Reveal a Role for RNA Processing Factor THRAP3 in the DNA Damage Response. Mol. Cell 2012, 46, 212–225. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.; Bacolla, A.; Del Mundo, I.M.; Zhao, J.; Wang, G.; Vasquez, K.M. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013, 41, 10345–10357. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, G.; d’Adda di Fagagna, F. Transcription and DNA Damage: Holding Hands or Crossing Swords? J. Mol. Biol. 2017, 429, 3215–3229. [Google Scholar] [CrossRef]
- Kakarougkas, A.; Downs, J.A.; Jeggo, P.A. The PBAF chromatin remodeling complex represses transcription and promotes rapid repair at DNA double-strand breaks. Mol. Cell Oncol 2015, 2, e970072. [Google Scholar] [CrossRef]
- Ui, A.; Nagaura, Y.; Yasui, A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol. Cell 2015, 58, 468–482. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Yang, H.; Wang, H. The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage. J. Biol. Chem. 2014, 289, 32883–32894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alagoz, M.; Katsuki, Y.; Ogiwara, H.; Ogi, T.; Shibata, A.; Kakarougkas, A.; Jeggo, P. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015, 43, 7931–7944. [Google Scholar] [CrossRef] [Green Version]
- Campbell, S.; Ismail, I.H.; Young, L.C.; Poirier, G.G.; Hendzel, M.J. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 2013, 12, 2675–2683. [Google Scholar] [CrossRef]
- Ismail, I.H.; Gagne, J.P.; Caron, M.C.; McDonald, D.; Xu, Z.; Masson, J.Y.; Poirier, G.G.; Hendzel, M.J. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012, 40, 5497–5510. [Google Scholar] [CrossRef] [Green Version]
- Chiarle, R.; Zhang, Y.; Frock, R.L.; Lewis, S.M.; Molinie, B.; Ho, Y.J.; Myers, D.R.; Choi, V.W.; Compagno, M.; Malkin, D.J.; et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 2011, 147, 107–119. [Google Scholar] [CrossRef] [Green Version]
- Burman, B.; Zhang, Z.Z.; Pegoraro, G.; Lieb, J.D.; Misteli, T. Histone modifications predispose genome regions to breakage and translocation. Genes Dev. 2015. [Google Scholar] [CrossRef] [Green Version]
- Keskin, H.; Shen, Y.; Huang, F.; Patel, M.; Yang, T.; Ashley, K.; Mazin, A.V.; Storici, F. Transcript-RNA-templated DNA recombination and repair. Nature 2014, 515, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Germain, D.R.; Poon, H.-Y.; Hildebrandt, M.R.; Monckton, E.A.; McDonald, D.; Hendzel, M.J.; Godbout, R. DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Mol. Cell. Biol. 2016, 36, 2794–2810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, C.; Pizzul, P.; Longhese, M.P.; Bonetti, D. Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front. Cell Dev. Biol. 2021, 8, 618157. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Wang, Y.; Qiao, J. PHF1 is required for chromosome alignment and asymmetric division during mouse meiotic oocyte maturation. Cell Cycle 2018, 17, 2447–2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Y.; Lu, D.; Jiang, H.; Chi, X.; Zhang, H. EZH2 is required for mouse oocyte meiotic maturation by interacting with and stabilizing spindle assembly checkpoint protein BubRI. Nucleic Acids Res. 2016, 44, 7659–7672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertolini, G.; Roz, L.; Perego, P.; Tortoreto, M.; Fontanella, E.; Gatti, L.; Pratesi, G.; Fabbri, A.; Andriani, F.; Tinelli, S.; et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc. Natl. Acad. Sci. USA 2009, 106, 16281–16286. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.M.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef] [Green Version]
- Song, L.B.; Zeng, M.S.; Liao, W.T.; Zhang, L.; Mo, H.Y.; Liu, W.L.; Shao, J.Y.; Wu, Q.L.; Li, M.Z.; Xia, Y.F.; et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006, 66, 6225–6232. [Google Scholar] [CrossRef] [Green Version]
- Song, L.B.; Li, J.; Liao, W.T.; Feng, Y.; Yu, C.P.; Hu, L.J.; Kong, Q.L.; Xu, L.H.; Zhang, X.; Liu, W.L.; et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J. Clin. Invest. 2009, 119, 3626–3636. [Google Scholar] [CrossRef] [Green Version]
- Oishi, N.; Wang, X.W. Novel therapeutic strategies for targeting liver cancer stem cells. Int. J. Biol. Sci. 2011, 7, 517–535. [Google Scholar] [CrossRef] [Green Version]
- Wiederschain, D.; Chen, L.; Johnson, B.; Bettano, K.; Jackson, D.; Taraszka, J.; Wang, Y.K.; Jones, M.D.; Morrissey, M.; Deeds, J.; et al. Contribution of Polycomb Homologues Bmi-1 and Mel-18 to Medulloblastoma Pathogenesis. Mol. Cell. Biol. 2007, 27, 4968–4979. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Hu, B.; Li, T.; Ma, J.; Alam, G.; Gunning, W.T.; Ding, H.F. Bmi-1 is essential for the tumorigenicity of neuroblastoma cells. Am. J. Pathol. 2007, 170, 1370–1378. [Google Scholar] [CrossRef] [Green Version]
- Molofsky, A.V.; He, S.; Bydon, M.; Morrison, S.J.; Pardal, R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19 Arf senescence pathways. Genes Dev. 2005, 19, 1432–1437. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, J.L.; Kieboom, K.; Marino, S.; DePinho, R.A.; Van Lohuizen, M. The oncogene and Polycombgroup gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999, 397, 164–168. [Google Scholar] [CrossRef]
- Bommi, P.V.; Dimri, M.; Sahasrabuddhe, A.A.; Khandekar, J.D.; Dimri, G.P. The polycomb group protein BMI1 is a transcriptional target of HDAC inhibitors. Cell Cycle 2010, 9, 2663–2673. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.W.; Lee, S.; Seo, M.S.; Park, S.B.; Kurtz, A.; Kang, S.K.; Kang, K.S. Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell. Mol. Life Sci. 2010, 67, 1165–1176. [Google Scholar] [CrossRef] [Green Version]
- Kaneta, Y.; Arai, M.A.; Ishikawa, N.; Toume, K.; Koyano, T.; Kowithayakorn, T.; Chiba, T.; Iwama, A.; Ishibashi, M. Identification of BMI1 Promoter Inhibitors from Beaumontia murtonii and Eugenia operculata. J. Nat. Prod. 2017, 80, 1853–1859. [Google Scholar] [CrossRef]
- Bansal, N.; Bartucci, M.; Yusuff, S.; Davis, S.; Flaherty, K.; Huselid, E.; Patrizii, M.; Jones, D.; Cao, L.; Sydorenko, N.; et al. BMI-1 targeting interferes with patient-derived tumor-initiating cell survival and tumor growth in prostate cancer. Clin. Cancer Res. 2016, 22, 6176–6191. [Google Scholar] [CrossRef] [Green Version]
- Bartucci, M.; Hussein, M.S.; Huselid, E.; Flaherty, K.; Patrizii, M.; Laddha, S.V.; Kui, C.; Bigos, R.A.; Gilleran, J.A.; El Ansary, M.M.S.; et al. Synthesis and Characterization of Novel BMI1 Inhibitors Targeting Cellular Self-Renewal in Hepatocellular Carcinoma. Target. Oncol. 2017, 12, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Strebhardt, K. Multifaceted polo-like kinases: Drug targets and antitargets for cancer therapy. Nat. Rev. Drug Discov. 2010, 9, 643–660. [Google Scholar] [CrossRef]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 2006, 6, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Dimri, M.; Cho, J.H.; Kang, M.; Dimri, G.P. PLK1 inhibition down-regulates polycomb group protein BMI1 via modulation of the miR-200c/141 cluster. J. Biol. Chem. 2015, 290, 3033–3044. [Google Scholar] [CrossRef] [Green Version]
- Voncken, J.W.; Schweizer, D.; Aagaard, L.; Sattler, L.; Jantsch, M.F.; Van Lohuizen, M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J. Cell Sci. 1999, 112, 4627–4639. [Google Scholar]
- Voncken, J.W.; Niessen, H.; Neufeld, B.; Rennefahrt, U.; Dahlmans, V.; Kubben, N.; Holzer, B.; Ludwig, S.; Rapp, U.R. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J. Biol. Chem. 2005, 280, 5178–5187. [Google Scholar] [CrossRef] [Green Version]
- Kreso, A.; van Galen, P.; Pedley, N.M.; Lima-Fernandes, E.; Frelin, C.; Davis, T.; Cao, L.; Baiazitov, R.; Du, W.; Sydorenko, N.; et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 2014, 20, 29–36. [Google Scholar] [CrossRef]
- Yong, K.J.; Basseres, D.S.; Welner, R.S.; Zhang, W.C.; Yang, H.; Yan, B.; Alberich-Jorda, M.; Zhang, J.; De Figueiredo-Pontes, L.L.; Battelli, C.; et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBPα expression. Sci. Transl. Med. 2016, 8, 350ra104. [Google Scholar] [CrossRef] [Green Version]
- Bolomsky, A.; Schlangen, K.; Schreiner, W.; Zojer, N.; Ludwig, H. Targeting of BMI-1 with PTC-209 shows potent anti-myeloma activity and impairs the tumour microenvironment. J. Hematol. Oncol. 2016, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Sahasrabuddhe, A.A.; Dimri, M.; Bommi, P.V.; Dimri, G.P. βTrCP regulates BMI1 protein turnover via ubiquitination and degradation. Cell Cycle 2011, 10, 1322–1330. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fitieh, A.; Locke, A.J.; Motamedi, M.; Ismail, I.H. The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability. Int. J. Mol. Sci. 2021, 22, 2976. https://doi.org/10.3390/ijms22062976
Fitieh A, Locke AJ, Motamedi M, Ismail IH. The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability. International Journal of Molecular Sciences. 2021; 22(6):2976. https://doi.org/10.3390/ijms22062976
Chicago/Turabian StyleFitieh, Amira, Andrew J. Locke, Mobina Motamedi, and Ismail Hassan Ismail. 2021. "The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability" International Journal of Molecular Sciences 22, no. 6: 2976. https://doi.org/10.3390/ijms22062976





