Implications of the Wilms’ Tumor Suppressor Wt1 in Cardiomyocyte Differentiation
Abstract
:1. Introduction
2. Results
2.1. Wt1 Expression in Developing and Adult Hearts
2.2. Wt1 Expression in Infarcted Hearts
2.3. Cardiomyocyte Differentiation In Vitro
2.4. Wt1 Overexpression Affects the Course of Cardiomyocyte Differentiation
3. Discussion
4. Materials and Methods
4.1. Mice and Tissue Preparation
4.2. Cell Culture
4.2.1. Mouse Embryonic Stem Cell Culture
4.2.2. mESC Differentiation by the Hanging Drop Method
4.3. Electroporation
4.4. Quantitative RT-PCR
4.5. Mouse Tissue Samples, Histology and Immunohistology
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMEM | Dulbecco’s Modified Eagle Medium |
| E | Embryonic day |
| EBs | Embryoid bodies |
| EPDC | Epicardial-derived cells |
| FBS | Fetal bovine serum |
| LAD | Left anterior descending coronary artery |
| LIF | Leukemia inhibitory factor |
| MEF | Mouse embryonic fibroblasts |
| mESCs | Mouse embryonic stem cells |
| MI | Myocardial infarction |
| P | Postnatal day |
| pt | Post transfection |
| qRT-PCR | Quantitative reverse-transcription polymerase chain reaction |
| S.E.M. | Standard error of the mean |
| Wt1 | Wilms’ tumor Suppressor 1 |
References
- Rackley, R.R.; Flenniken, A.M.; Kuriyan, N.P.; Kessler, P.M.; Stoler, M.H.; Williams, B.R. Expression of the Wilms’ tumor suppressor gene WT1 during mouse embryogenesis. Cell Growth Differ. 1993, 4, 1023–1031. [Google Scholar]
- Hastie, N.D. Wilms’ tumour 1 (WT1) in development, homeostasis and disease. Development 2017, 144, 2862–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, K.D.; Wagner, N.; Schedl, A. The complex life of WT1. J. Cell Sci. 2003, 116, 1653–1658. [Google Scholar] [CrossRef] [Green Version]
- Hohenstein, P.; Hastie, N.D. The many facets of the Wilms’ tumour gene, WT1. Hum. Mol. Genet. 2006, 15, R196–R201. [Google Scholar] [CrossRef]
- Oji, Y.; Miyoshi, S.; Maeda, H.; Hayashi, S.; Tamaki, H.; Nakatsuka, S.; Yao, M.; Takahashi, E.; Nakano, Y.; Hirabayashi, H.; et al. Overexpression of the Wilms’ tumor gene WT1 in de novo lung cancers. Int. J. Cancer 2002, 100, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Rampal, R.; Figueroa, M.E. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica 2016, 101, 672–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huff, V.; Miwa, H.; Haber, D.A.; Call, K.M.; Housman, D.; Strong, L.C.; Saunders, G.F. Evidence for WT1 as a Wilms tumor (WT) gene: Intragenic germinal deletion in bilateral WT. Am. J. Hum. Genet. 1991, 48, 997–1003. [Google Scholar]
- Haber, D.A.; Buckler, A.J.; Glaser, T.; Call, K.M.; Pelletier, J.; Sohn, R.L.; Douglass, E.C.; Housman, D.E. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumor. Cell 1990, 61, 1257–1269. [Google Scholar] [CrossRef]
- Wagner, K.D.; Cherfils-Vicini, J.; Hosen, N.; Hohenstein, P.; Gilson, E.; Hastie, N.D.; Michiels, J.F.; Wagner, N. The Wilms’ tumour suppressor Wt1 is a major regulator of tumour angiogenesis and progression. Nat. Commun. 2014, 5, 5852. [Google Scholar] [CrossRef] [Green Version]
- Belali, T.; Wodi, C.; Clark, B.; Cheung, M.K.; Craig, T.J.; Wheway, G.; Wagner, N.; Wagner, K.D.; Roberts, S.; Porazinski, S.; et al. WT1 activates transcription of the splice factor kinase SRPK1 gene in PC3 and K562 cancer cells in the absence of corepressor BASP1. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194642. [Google Scholar] [CrossRef]
- Wagner, K.D.; El Maï, M.; Ladomery, M.; Belali, T.; Leccia, N.; Michiels, J.F.; Wagner, N. Altered VEGF Splicing Isoform Balance in Tumor Endothelium Involves Activation of Splicing Factors Srpk1 and Srsf1 by the Wilms’ Tumor Suppressor Wt1. Cells 2019, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Wagner, N.; Michiels, J.F.; Schedl, A.; Wagner, K.D. The Wilms’ tumour suppressor WT1 is involved in endothelial cell proliferation and migration: Expression in tumour vessels in vivo. Oncogene 2008, 27, 3662–3672. [Google Scholar] [CrossRef] [Green Version]
- Eisermann, K.; Tandon, S.; Bazarov, A.; Brett, A.; Fraizer, G.; Piontkivska, H. Evolutionary conservation of zinc finger transcription factor binding sites in promoters of genes co-expressed with WT1 in prostate cancer. BMC Genom. 2008, 9, 337. [Google Scholar] [CrossRef] [Green Version]
- Rauscher, F.J.; Morris, J.F.; Tournay, O.E.; Cook, D.M.; Curran, T. Binding of the Wilms’ tumor locus zinc finger protein to the EGR-1 consensus sequence. Science 1990, 250, 1259–1262. [Google Scholar] [CrossRef]
- Haber, D.A.; Sohn, R.L.; Buckler, A.J.; Pelletier, J.; Call, K.M.; Housman, D.E. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc. Natl. Acad. Sci. USA 1991, 88, 9618–9622. [Google Scholar] [CrossRef] [Green Version]
- Toska, E.; Roberts, S.G. Mechanisms of transcriptional regulation by WT1 (Wilms’ tumour 1). Biochem. J. 2014, 461, 15–32. [Google Scholar] [CrossRef]
- Laity, J.H.; Dyson, H.J.; Wright, P.E. Molecular basis for modulation of biological function by alternate splicing of the Wilms’ tumor suppressor protein. Proc. Natl. Acad. Sci. USA 2000, 97, 11932–11935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, S.H.; Charlieu, J.P.; Miyagawa, K.; Engelkamp, D.; Rassoulzadegan, M.; Ross, A.; Cuzin, F.; van Heyningen, V.; Hastie, N.D. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 1995, 81, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Alberta, J.A.; Springett, G.M.; Rayburn, H.; Natoli, T.A.; Loring, J.; Kreidberg, J.A.; Housman, D. Role of the WT1 tumor suppressor in murine hematopoiesis. Blood 2003, 101, 2570–2574. [Google Scholar] [CrossRef]
- Herzer, U.; Crocoll, A.; Barton, D.; Howells, N.; Englert, C. The Wilms tumor suppressor gene wt1 is required for development of the spleen. Curr. Biol. 1999, 9, 837–840. [Google Scholar] [CrossRef] [Green Version]
- Moore, A.W.; McInnes, L.; Kreidberg, J.; Hastie, N.D.; Schedl, A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999, 126, 1845–1857. [Google Scholar]
- Wagner, N.; Wagner, K.D.; Hammes, A.; Kirschner, K.M.; Vidal, V.P.; Schedl, A.; Scholz, H. A splice variant of the Wilms’ tumour suppressor Wt1 is required for normal development of the olfactory system. Development 2005, 132, 1327–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, N.; Wagner, K.D.; Theres, H.; Englert, C.; Schedl, A.; Scholz, H. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms’ tumor transcription factor Wt1. Genes Dev. 2005, 19, 2631–2642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, N.; Wagner, K.D.; Scholz, H.; Kirschner, K.M.; Schedl, A. Intermediate filament protein nestin is expressed in developing kidney and heart and might be regulated by the Wilms’ tumor suppressor Wt1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 291, R779–R787. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.D.; Wagner, N.; Vidal, V.P.; Schley, G.; Wilhelm, D.; Schedl, A.; Englert, C.; Scholz, H. The Wilms’ tumor gene Wt1 is required for normal development of the retina. EMBO J. 2002, 21, 1398–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnerwitzki, D.; Hayn, C.; Perner, B.; Englert, C. Wt1 Positive dB4 Neurons in the Hindbrain Are Crucial for Respiration. Front. Neurosci. 2020, 14, 529487. [Google Scholar] [CrossRef]
- Weiss, A.C.; Rivera-Reyes, R.; Englert, C.; Kispert, A. Expansion of the renal capsular stroma, ureteric bud branching defects and cryptorchidism in mice with Wilms tumor 1 gene deletion in the stromal compartment of the developing kidney. J. Pathol. 2020, 252, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Schnerwitzki, D.; Perry, S.; Ivanova, A.; Caixeta, F.V.; Cramer, P.; Günther, S.; Weber, K.; Tafreshiha, A.; Becker, L.; Vargas Panesso, I.L.; et al. Neuron-specific inactivation of. Life Sci. Alliance 2018, 1, e201800106. [Google Scholar] [CrossRef] [Green Version]
- Nathan, A.; Reinhardt, P.; Kruspe, D.; Jörß, T.; Groth, M.; Nolte, H.; Habenicht, A.; Herrmann, J.; Holschbach, V.; Toth, B.; et al. The Wilms tumor protein Wt1 contributes to female fertility by regulating oviductal proteostasis. Hum. Mol. Genet. 2017, 26, 1694–1705. [Google Scholar] [CrossRef] [Green Version]
- Bharathavikru, R.; Dudnakova, T.; Aitken, S.; Slight, J.; Artibani, M.; Hohenstein, P.; Tollervey, D.; Hastie, N. Transcription factor Wilms’ tumor 1 regulates developmental RNAs through 3′ UTR interaction. Genes Dev. 2017, 31, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, S.; Ho, J.; Pandey, P.; Macisaac, K.; Taglienti, M.; Xiang, M.; Alterovitz, G.; Ramoni, M.; Fraenkel, E.; Kreidberg, J.A. Genomic characterization of Wilms’ tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development 2010, 137, 1189–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirschner, K.M.; Wagner, N.; Wagner, K.D.; Wellmann, S.; Scholz, H. The Wilms tumor suppressor Wt1 promotes cell adhesion through transcriptional activation of the alpha4integrin gene. J. Biol. Chem. 2006, 281, 31930–31939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreidberg, J.A.; Sariola, H.; Loring, J.M.; Maeda, M.; Pelletier, J.; Housman, D.; Jaenisch, R. WT-1 is required for early kidney development. Cell 1993, 74, 679–691. [Google Scholar] [CrossRef]
- Lee, S.B.; Huang, K.; Palmer, R.; Truong, V.B.; Herzlinger, D.; Kolquist, K.A.; Wong, J.; Paulding, C.; Yoon, S.K.; Gerald, W.; et al. The Wilms tumor suppressor WT1 encodes a transcriptional activator of amphiregulin. Cell 1999, 98, 663–673. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Estrada, O.M.; Lettice, L.A.; Essafi, A.; Guadix, J.A.; Slight, J.; Velecela, V.; Hall, E.; Reichmann, J.; Devenney, P.S.; Hohenstein, P.; et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 2010, 42, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Miquerol, L.; Kelly, R.G. Organogenesis of the vertebrate heart. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.D.; Wagner, N.; Bondke, A.; Nafz, B.; Flemming, B.; Theres, H.; Scholz, H. The Wilms’ tumor suppressor Wt1 is expressed in the coronary vasculature after myocardial infarction. FASEB J. 2002, 16, 1117–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duim, S.N.; Kurakula, K.; Goumans, M.J.; Kruithof, B.P. Cardiac endothelial cells express Wilms’ tumor-1: Wt1 expression in the developing, adult and infarcted heart. J. Mol. Cell. Cardiol. 2015, 81, 127–135. [Google Scholar] [CrossRef]
- Männer, J.; Wessel, A.; Yelbuz, T.M. How does the tubular embryonic heart work? Looking for the physical mechanism generating unidirectional blood flow in the valveless embryonic heart tube. Dev. Dyn. 2010, 239, 1035–1046. [Google Scholar] [CrossRef]
- Armstrong, J.F.; Pritchard-Jones, K.; Bickmore, W.A.; Hastie, N.D.; Bard, J.B. The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 1993, 40, 85–97. [Google Scholar] [CrossRef]
- Vicente-Steijn, R.; Scherptong, R.W.; Kruithof, B.P.; Duim, S.N.; Goumans, M.J.; Wisse, L.J.; Zhou, B.; Pu, W.T.; Poelmann, R.E.; Schalij, M.J.; et al. Regional differences in WT-1 and Tcf21 expression during ventricular development: Implications for myocardial compaction. PLoS ONE 2015, 10, e0136025. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Ma, Q.; Rajagopal, S.; Wu, S.M.; Domian, I.; Rivera-Feliciano, J.; Jiang, D.; von Gise, A.; Ikeda, S.; Chien, K.R.; et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008, 454, 109–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Smith, C.L.; Hall, J.A.; Lee, I.; Luby-Phelps, K.; Tallquist, M.D. Epicardial spindle orientation controls cell entry into the myocardium. Dev. Cell 2010, 19, 114–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, C.L.; Martin, J.C.; Sun, Y.; Cui, L.; Wang, L.; Ouyang, K.; Yang, L.; Bu, L.; Liang, X.; Zhang, X.; et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008, 454, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velecela, V.; Torres-Cano, A.; García-Melero, A.; Ramiro-Pareta, M.; Müller-Sánchez, C.; Segarra-Mondejar, M.; Chau, Y.Y.; Campos-Bonilla, B.; Reina, M.; Soriano, F.X.; et al. Epicardial cell shape and maturation are regulated by Wt1 via transcriptional control of. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Sereti, K.I.; Nguyen, N.B.; Kamran, P.; Zhao, P.; Ranjbarvaziri, S.; Park, S.; Sabri, S.; Engel, J.L.; Sung, K.; Kulkarni, R.P.; et al. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat. Commun. 2018, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Cano, E.; Carmona, R.; Ruiz-Villalba, A.; Rojas, A.; Chau, Y.Y.; Wagner, K.D.; Wagner, N.; Hastie, N.D.; Muñoz-Chápuli, R.; Pérez-Pomares, J.M. Extracardiac septum transversum/proepicardial endothelial cells pattern embryonic coronary arterio-venous connections. Proc. Natl. Acad. Sci. USA 2016, 113, 656–661. [Google Scholar] [CrossRef] [Green Version]
- Wagner, N.; Morrison, H.; Pagnotta, S.; Michiels, J.F.; Schwab, Y.; Tryggvason, K.; Schedl, A.; Wagner, K.D. The podocyte protein nephrin is required for cardiac vessel formation. Hum. Mol. Genet. 2011, 20, 2182–2194. [Google Scholar] [CrossRef] [Green Version]
- Wagner, K.D.; Wagner, N.; Wellmann, S.; Schley, G.; Bondke, A.; Theres, H.; Scholz, H. Oxygen-regulated expression of the Wilms’ tumor suppressor Wt1 involves hypoxia-inducible factor-1 (HIF-1). FASEB J. 2003, 17, 1364–1366. [Google Scholar] [CrossRef] [Green Version]
- Carmona, R.; Barrena, S.; López Gambero, A.J.; Rojas, A.; Muñoz-Chápuli, R. Epicardial cell lineages and the origin of the coronary endothelium. FASEB J. 2020, 34, 5223–5239. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, N.; Nakazawa, N.; Kurata, Y.; Yaura, H.; Taufiq, F.; Minato, H.; Yoshida, A.; Ninomiya, H.; Nakayama, Y.; Kuwabara, M.; et al. Tbx18-positive cells differentiated from murine ES cells serve as proepicardial progenitors to give rise to vascular smooth muscle cells and fibroblasts. Biomed. Res. 2017, 38, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Smart, N.; Bollini, S.; Dubé, K.N.; Vieira, J.M.; Zhou, B.; Davidson, S.; Yellon, D.; Riegler, J.; Price, A.N.; Lythgoe, M.F.; et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature 2011, 474, 640–644. [Google Scholar] [CrossRef] [Green Version]
- Von Gise, A.; Zhou, B.; Honor, L.B.; Ma, Q.; Petryk, A.; Pu, W.T. WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev. Biol. 2011, 356, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Wilm, B.; Ipenberg, A.; Hastie, N.D.; Burch, J.B.; Bader, D.M. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 2005, 132, 5317–5328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudat, C.; Kispert, A. Wt1 and epicardial fate mapping. Circ. Res. 2012, 111, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoffels, V.M.; Grieskamp, T.; Norden, J.; Mommersteeg, M.T.; Rudat, C.; Kispert, A. Tbx18 and the fate of epicardial progenitors. Nature 2009, 458, E8–E9. [Google Scholar] [CrossRef] [PubMed]
- Maltsev, V.A.; Rohwedel, J.; Hescheler, J.; Wobus, A.M. Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech. Dev. 1993, 44, 41–50. [Google Scholar] [CrossRef]
- Boheler, K.R.; Czyz, J.; Tweedie, D.; Yang, H.T.; Anisimov, S.V.; Wobus, A.M. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 2002, 91, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Varlakhanova, N.V.; Cotterman, R.F.; de Vries, W.N.; Morgan, J.; Donahue, L.R.; Murray, S.; Knowles, B.B.; Knoepfler, P.S. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 2010, 80, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Soonpaa, M.H.; Adler, E.D.; Roepke, T.K.; Kattman, S.J.; Kennedy, M.; Henckaerts, E.; Bonham, K.; Abbott, G.W.; Linden, R.M.; et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 2008, 453, 524–528. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, Y.H.; Lee, Y.A.; Jung, S.E.; Hong, Y.H.; Lee, E.J.; Kim, B.G.; Hwang, S.; Do, J.T.; Pang, M.G.; et al. Platelet-derived growth factor receptor-alpha positive cardiac progenitor cells derived from multipotent germline stem cells are capable of cardiomyogenesis in vitro and in vivo. Oncotarget 2017, 8, 29643–29656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meilhac, S.M.; Buckingham, M.E. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018, 15, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Wessels, A.; van den Hoff, M.J.; Adamo, R.F.; Phelps, A.L.; Lockhart, M.M.; Sauls, K.; Briggs, L.E.; Norris, R.A.; van Wijk, B.; Perez-Pomares, J.M.; et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev. Biol. 2012, 366, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Zeng, B.; Ren, X.F.; Cao, F.; Zhou, X.Y.; Zhang, J. Developmental patterns and characteristics of epicardial cell markers Tbx18 and Wt1 in murine embryonic heart. J. Biomed. Sci. 2011, 18, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redpath, A.N.; Smart, N. Recapturing embryonic potential in the adult epicardium: Prospects for cardiac repair. Stem Cells Transl. Med. 2021, 10, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Marín-Juez, R.; El-Sammak, H.; Helker, C.S.M.; Kamezaki, A.; Mullapuli, S.T.; Bibli, S.I.; Foglia, M.J.; Fleming, I.; Poss, K.D.; Stainier, D.Y.R. Coronary Revascularization During Heart Regeneration Is Regulated by Epicardial and Endocardial Cues and Forms a Scaffold for Cardiomyocyte Repopulation. Dev. Cell 2019, 51, 503–515. [Google Scholar] [CrossRef]
- Liu, Y.H.; Lai, L.P.; Huang, S.Y.; Lin, Y.S.; Wu, S.C.; Chou, C.J.; Lin, J.L. Developmental origin of postnatal cardiomyogenic progenitor cells. Future Sci. OA 2016, 2, FSO120. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.N.; Thatcher, J.E.; McAnally, J.; Kong, Y.; Qi, X.; Tan, W.; DiMaio, J.M.; Amatruda, J.F.; Gerard, R.D.; Hill, J.A.; et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science 2012, 338, 1599–1603. [Google Scholar] [CrossRef] [Green Version]
- Van Wijk, B.; Gunst, Q.D.; Moorman, A.F.; van den Hoff, M.J. Cardiac regeneration from activated epicardium. PLoS ONE 2012, 7, e44692. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Honor, L.B.; Ma, Q.; Oh, J.H.; Lin, R.Z.; Melero-Martin, J.M.; von Gise, A.; Zhou, P.; Hu, T.; He, L.; et al. Thymosin beta 4 treatment after myocardial infarction does not reprogram epicardial cells into cardiomyocytes. J. Mol. Cell. Cardiol. 2012, 52, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Bollini, S.; Vieira, J.M.; Howard, S.; Dubè, K.N.; Balmer, G.M.; Smart, N.; Riley, P.R. Re-activated adult epicardial progenitor cells are a heterogeneous population molecularly distinct from their embryonic counterparts. Stem Cells Dev. 2014, 23, 1719–1730. [Google Scholar] [CrossRef]
- Zhou, B.; Honor, L.B.; He, H.; Ma, Q.; Oh, J.H.; Butterfield, C.; Lin, R.Z.; Melero-Martin, J.M.; Dolmatova, E.; Duffy, H.S.; et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Investig. 2011, 121, 1894–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanek, K.; Torella, D.; Sheikh, F.; De Angelis, A.; Nurzynska, D.; Silvestri, F.; Beltrami, C.A.; Bussani, R.; Beltrami, A.P.; Quaini, F.; et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc. Natl. Acad. Sci. USA 2005, 102, 8692–8697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Krueger, K.; Catanese, L.; Sciesielski, L.K.; Kirschner, K.M.; Scholz, H. Deletion of an intronic HIF-2α binding site suppresses hypoxia-induced WT1 expression. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 71–83. [Google Scholar] [CrossRef]
- Kirschner, K.M.; Sciesielski, L.K.; Krueger, K.; Scholz, H. Wilms tumor protein-dependent transcription of VEGF receptor 2 and hypoxia regulate expression of the testis-promoting gene. J. Biol. Chem. 2017, 292, 20281–20291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Maï, M.; Wagner, K.D.; Michiels, J.F.; Ambrosetti, D.; Borderie, A.; Destree, S.; Renault, V.; Djerbi, N.; Giraud-Panis, M.J.; Gilson, E.; et al. The Telomeric Protein TRF2 Regulates Angiogenesis by Binding and Activating the PDGFRβ Promoter. Cell Rep. 2014, 9, 1047–1060. [Google Scholar] [CrossRef] [Green Version]
- McCarty, G.; Awad, O.; Loeb, D.M. WT1 protein directly regulates expression of vascular endothelial growth factor and is a mediator of tumor response to hypoxia. J. Biol. Chem. 2011, 286, 43634–43643. [Google Scholar] [CrossRef] [Green Version]
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.D.; Guerquin-Kern, J.L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013, 493, 433–436. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, P.C.; Segers, V.F.; Davis, M.E.; MacGillivray, C.; Gannon, J.; Molkentin, J.D.; Robbins, J.; Lee, R.T. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat. Med. 2007, 13, 970–974. [Google Scholar] [CrossRef]
- Scholz, H.; Kirschner, K.M. Oxygen-Dependent Gene Expression in Development and Cancer: Lessons Learned from the Wilms’ Tumor Gene, WT1. Front. Mol. Neurosci. 2011, 4, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, R.L.; Mazzei, L.; Manucha, W. Implications of the transcription factor WT1 linked to the pathologic cardiac remodeling post-myocardial infarction. Clin. Investig. Arterioscler. 2019, 31, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Balbi, C.; Lodder, K.; Costa, A.; Moimas, S.; Moccia, F.; van Herwaarden, T.; Rosti, V.; Campagnoli, F.; Palmeri, A.; De Biasio, P.; et al. Reactivating endogenous mechanisms of cardiac regeneration via paracrine boosting using the human amniotic fluid stem cell secretome. Int. J. Cardiol. 2019, 287, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sivakumaran, P.; Newcomb, A.E.; Hernandez, D.; Harris, N.; Khanabdali, R.; Liu, G.S.; Kelly, D.J.; Pébay, A.; Hewitt, A.W.; et al. Cardiac Repair With a Novel Population of Mesenchymal Stem Cells Resident in the Human Heart. Stem Cells 2015, 33, 3100–3113. [Google Scholar] [CrossRef]
- Quijada, P.; Trembley, M.A.; Small, E.M. The Role of the Epicardium During Heart Development and Repair. Circ. Res. 2020, 126, 377–394. [Google Scholar] [CrossRef]
- Tyser, R.C.V.; Ibarra-Soria, X.; McDole, K.; Arcot Jayaram, S.; Godwin, J.; van den Brand, T.A.H.; Miranda, A.M.A.; Scialdone, A.; Keller, P.J.; Marioni, J.C.; et al. Characterization of a common progenitor pool of the epicardium and myocardium. Science 2021, 371. [Google Scholar] [CrossRef] [PubMed]
- Hescheler, J.; Fleischmann, B.K.; Lentini, S.; Maltsev, V.A.; Rohwedel, J.; Wobus, A.M.; Addicks, K. Embryonic stem cells: A model to study structural and functional properties in cardiomyogenesis. Cardiovasc. Res. 1997, 36, 149–162. [Google Scholar] [CrossRef]
- Maltsev, V.A.; Wobus, A.M.; Rohwedel, J.; Bader, M.; Hescheler, J. Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ. Res. 1994, 75, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Whitmill, A.; Liu, Y.; Timani, K.A.; Niu, Y.; He, J.J. Tip110 Deletion Impaired Embryonic and Stem Cell Development Involving Downregulation of Stem Cell Factors Nanog, Oct4, and Sox2. Stem Cells 2017, 35, 1674–1686. [Google Scholar] [CrossRef] [Green Version]
- Tallini, Y.N.; Greene, K.S.; Craven, M.; Spealman, A.; Breitbach, M.; Smith, J.; Fisher, P.J.; Steffey, M.; Hesse, M.; Doran, R.M.; et al. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc. Natl. Acad. Sci. USA 2009, 106, 1808–1813. [Google Scholar] [CrossRef] [Green Version]
- Noseda, M.; Harada, M.; McSweeney, S.; Leja, T.; Belian, E.; Stuckey, D.J.; Abreu Paiva, M.S.; Habib, J.; Macaulay, I.; de Smith, A.J.; et al. PDGFRα demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat. Commun. 2015, 6, 6930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Laake, L.W.; Passier, R.; Monshouwer-Kloots, J.; Nederhoff, M.G.; Ward-van Oostwaard, D.; Field, L.J.; van Echteld, C.J.; Doevendans, P.A.; Mummery, C.L. Monitoring of cell therapy and assessment of cardiac function using magnetic resonance imaging in a mouse model of myocardial infarction. Nat. Protoc. 2007, 2, 2551–2567. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, P. In vitro differentiation of mouse embryonic stem (mES) cells using the hanging drop method. J. Vis. Exp. 2008. [Google Scholar] [CrossRef] [Green Version]
- Wagner, K.D.; Du, S.; Martin, L.; Leccia, N.; Michiels, J.F.; Wagner, N. Vascular PPARβ/δ Promotes Tumor Angiogenesis and Progression. Cells 2019, 8, 1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keber, R.; Motaln, H.; Wagner, K.D.; Debeljak, N.; Rassoulzadegan, M.; Ačimovič, J.; Rozman, D.; Horvat, S. Mouse knockout of the cholesterogenic cytochrome P450 lanosterol 14alpha-demethylase (Cyp51) resembles Antley-Bixler syndrome. J. Biol. Chem. 2011, 286, 29086–29097. [Google Scholar] [CrossRef] [PubMed] [Green Version]


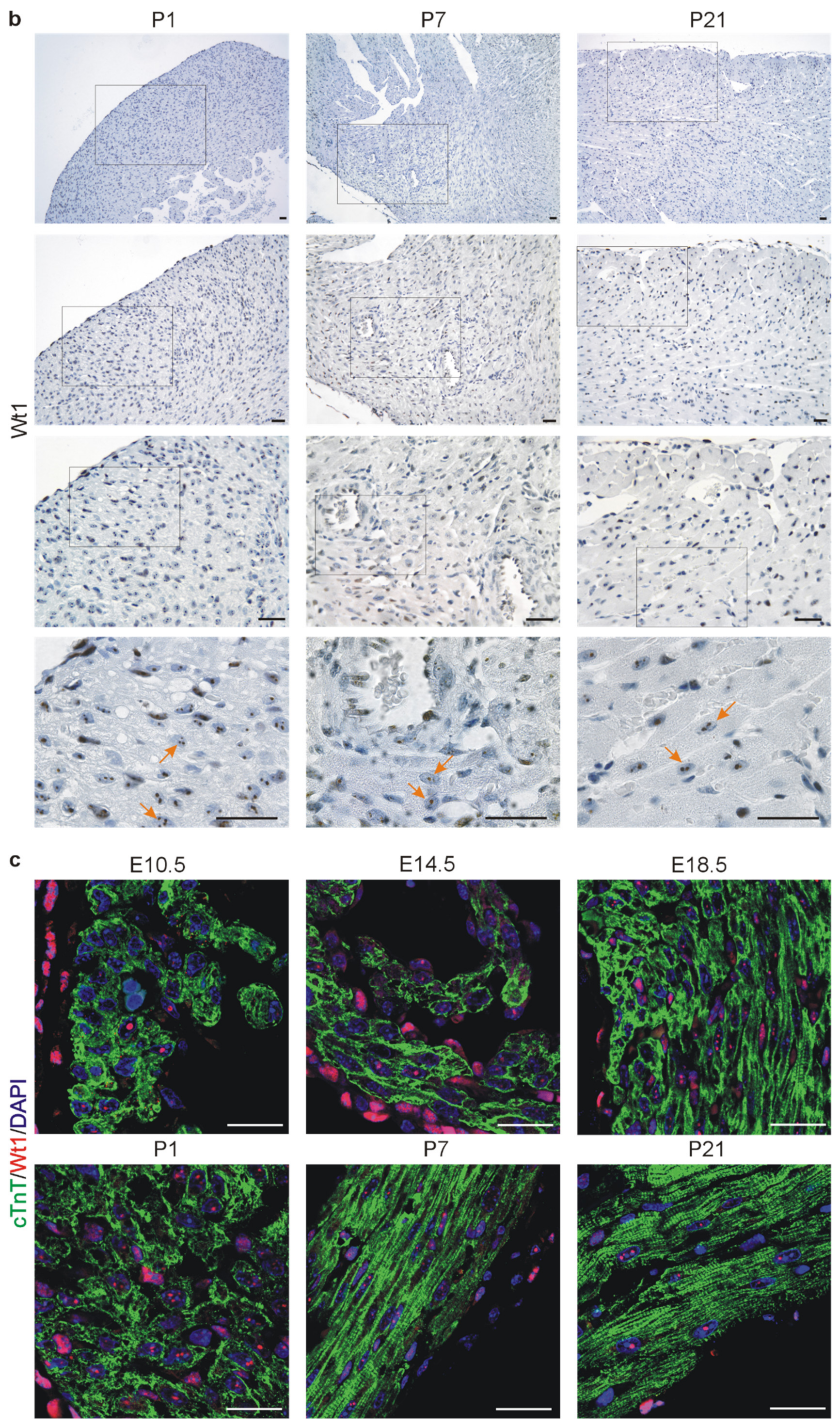
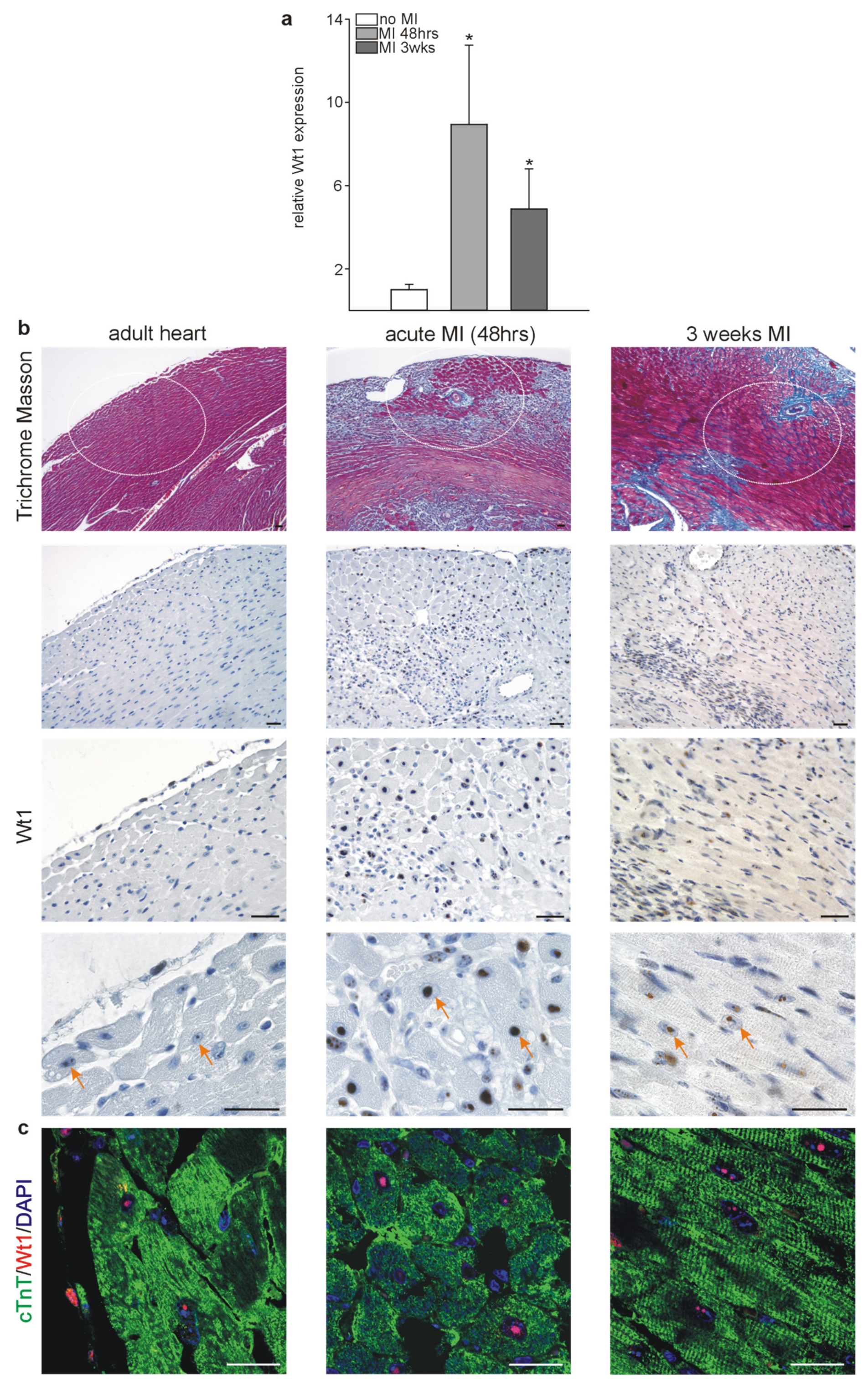
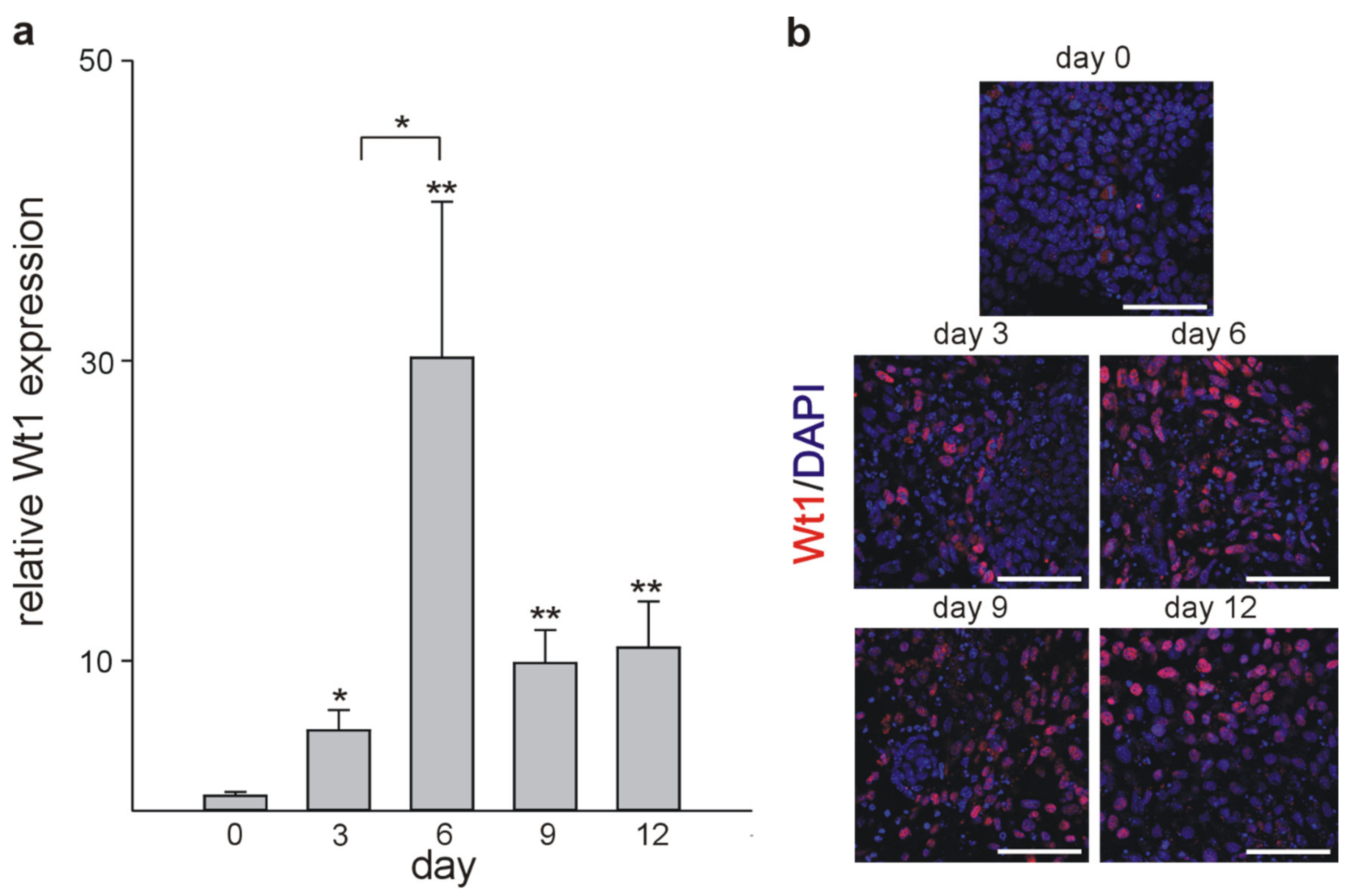


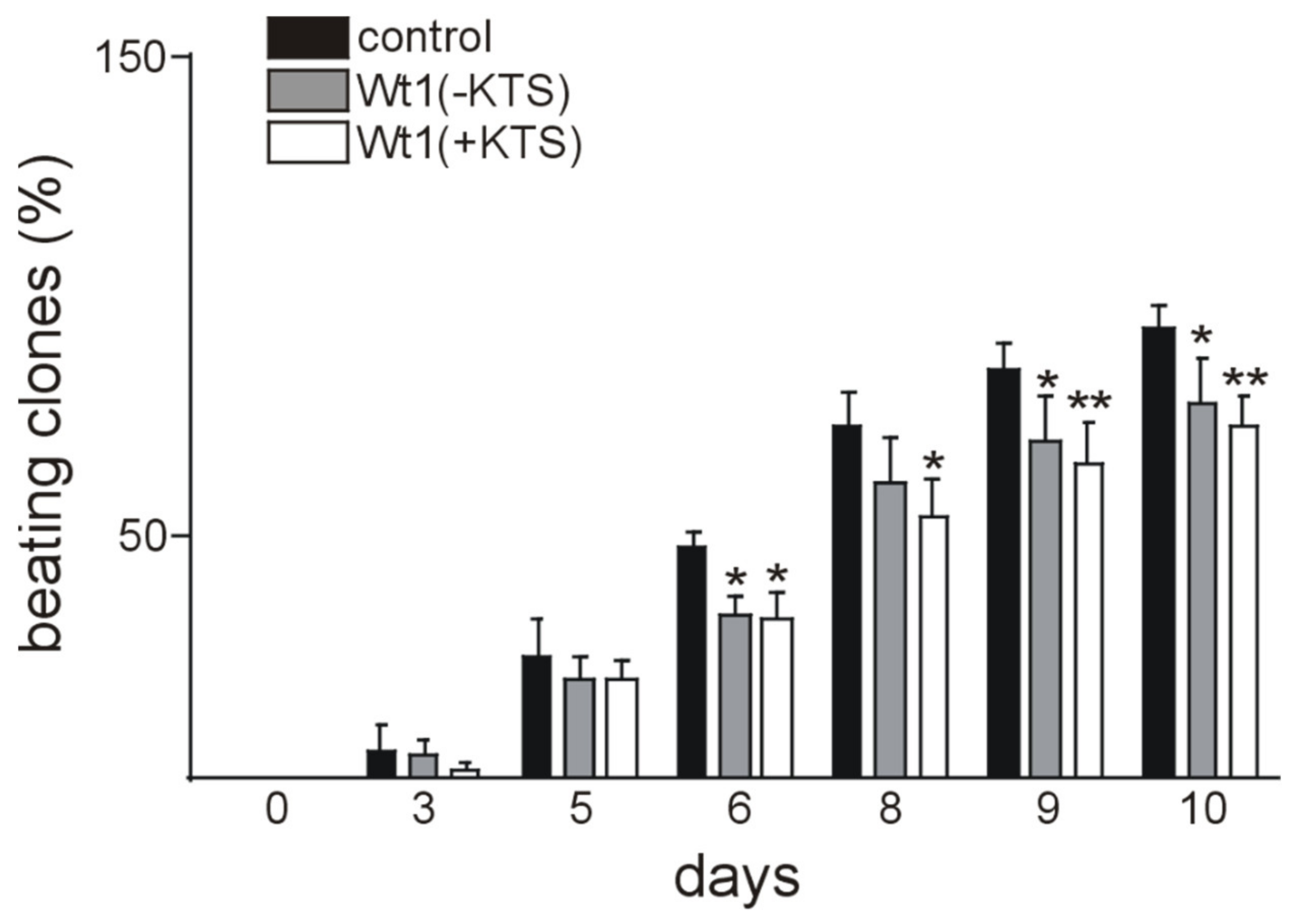
| Name | Sequence |
|---|---|
| Wt1 forward | CCA GCT CAG TGA AAT GGA CA [11] |
| Wt1 reverse | CTG TAC TGG GCA CCA CAG AG [11] |
| Kit forward | GCC TGA CGT GCA TTG ATC C [94] |
| Kit reverse | AGT GGC CTC GGC TTT TTC C [94] |
| Sox2 forward | CGC CCA GTA GAC TGC ACA |
| Sox2 reverse | CCC TCA CAT GTG CGA CAG |
| Oct4 forward | TGG GCG TTC TCT TTG GAA |
| Oct4 reverse | GTT GTC GGC TTC CTC CAC |
| Nanog forward | CAG GTT TCA GAA GCA GAA GTA CC |
| Nanog reverse | GGT TTT GAA ACC AGG TCT TAA CC |
| Myh6 forward | CCA AGA CTG TCC GGA ATG A |
| Myh6 reverse | TCC AAA GTG GAT CCT GAT GA |
| Myh7 forward | GCC TCC ATT GAT GAC TCT G |
| Myh7 reverse | CGC CTG TCA GCT TGT AAA TG |
| Nkx2–5 forward | ATT TTA CCC GGG AGC CTA CG |
| Nkx2–5 reverse | CAG CGC GCA CAG CTC TTT T |
| Kdr forward | AGT GGT ACT GGC AGC TAG AAG [94] |
| Kdr reverse | ACA AGC ATA CGG GCT TGT TT [94] |
| Pdgfra forward | ATG AGA GTG AGA TCG AAG GCA [94] |
| Pdgfra reverse | CGG CAA GGT ATG ATG GCA GAG [94] |
| Rplp0 forward | CAC TGG TCT AGG ACC CGA GAA G [95] |
| Rplp0 reverse | GGT GCC TCT GGA GAT TTT CG [95] |
| Gapdh forward | CCA ATG TGT CCG TCG TGG ATC T [48,95] |
| Gapdh reverse | GTT GAA GTC GCA GGA GAC AAC C [48,95] |
| Actb forward | CTT CCT CCC TGG AGA AGA GC [48,95] |
| Actb reverse | ATG CCA CAG GAT TCC ATA CC [48,95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, N.; Ninkov, M.; Vukolic, A.; Cubukcuoglu Deniz, G.; Rassoulzadegan, M.; Michiels, J.-F.; Wagner, K.-D. Implications of the Wilms’ Tumor Suppressor Wt1 in Cardiomyocyte Differentiation. Int. J. Mol. Sci. 2021, 22, 4346. https://doi.org/10.3390/ijms22094346
Wagner N, Ninkov M, Vukolic A, Cubukcuoglu Deniz G, Rassoulzadegan M, Michiels J-F, Wagner K-D. Implications of the Wilms’ Tumor Suppressor Wt1 in Cardiomyocyte Differentiation. International Journal of Molecular Sciences. 2021; 22(9):4346. https://doi.org/10.3390/ijms22094346
Chicago/Turabian StyleWagner, Nicole, Marina Ninkov, Ana Vukolic, Günseli Cubukcuoglu Deniz, Minoo Rassoulzadegan, Jean-François Michiels, and Kay-Dietrich Wagner. 2021. "Implications of the Wilms’ Tumor Suppressor Wt1 in Cardiomyocyte Differentiation" International Journal of Molecular Sciences 22, no. 9: 4346. https://doi.org/10.3390/ijms22094346








