The Beneficial Effects of Combining Anti-Aβ Antibody NP106 and Curcumin Analog TML-6 on the Treatment of Alzheimer’s Disease in APP/PS1 Mice
Abstract
:1. Introduction
2. Results
2.1. Combination Treatment Outperformed NP106 or TML-6 Monotherapy in Reducing Aβ Levels in the Brain of APP/PS1 Mice
2.2. Effects of Combination Treatment on the Nesting Behavioral Test
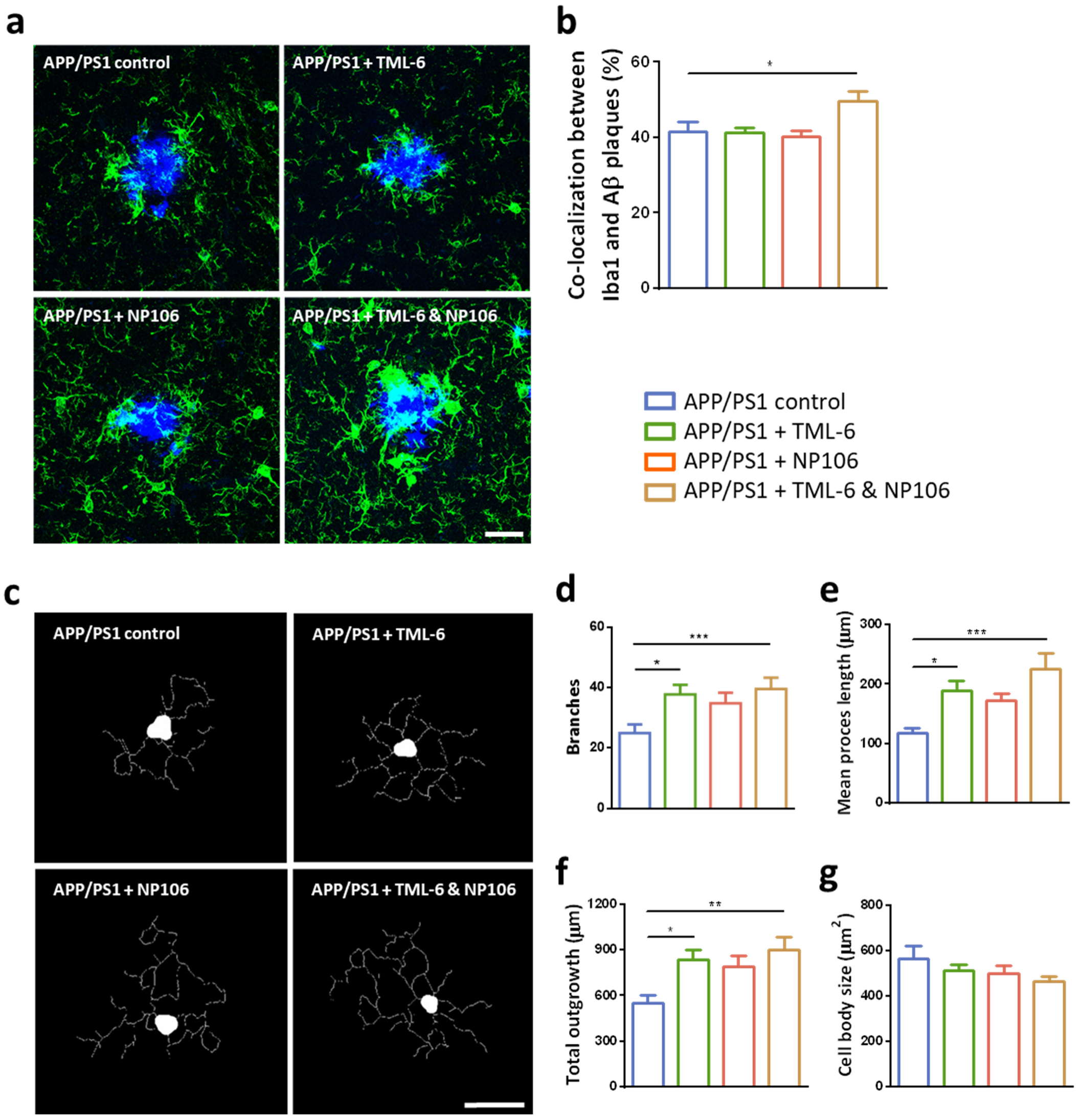
2.3. Effects of Combination Treatment on Microglial Aβ Phagocytosis and the Morphological Changes of Reactive Microglia
2.4. Anti-Inflammatory Effects of Combination Treatment on the Reduction of Cerebral Proinflammatory Cytokines
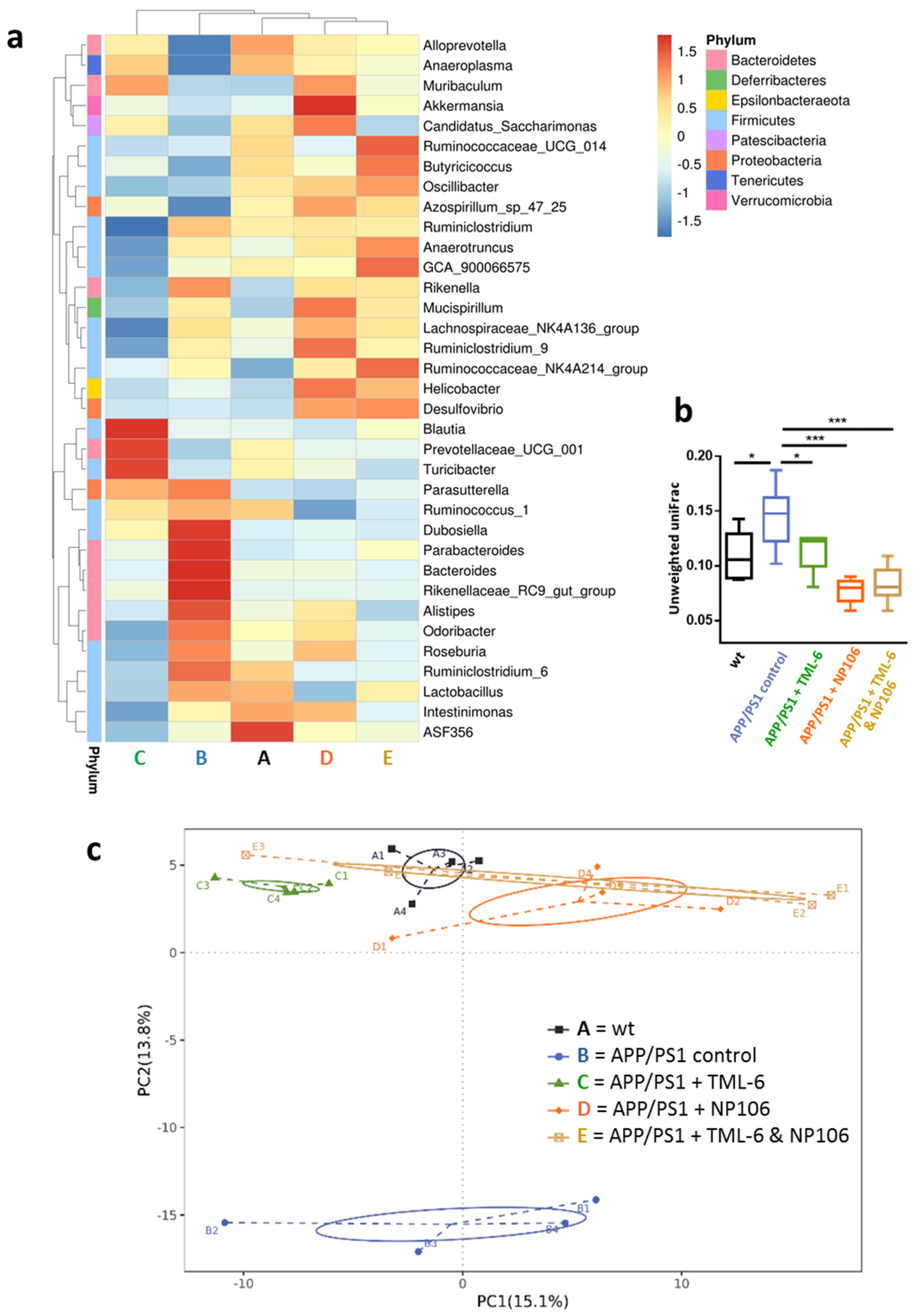
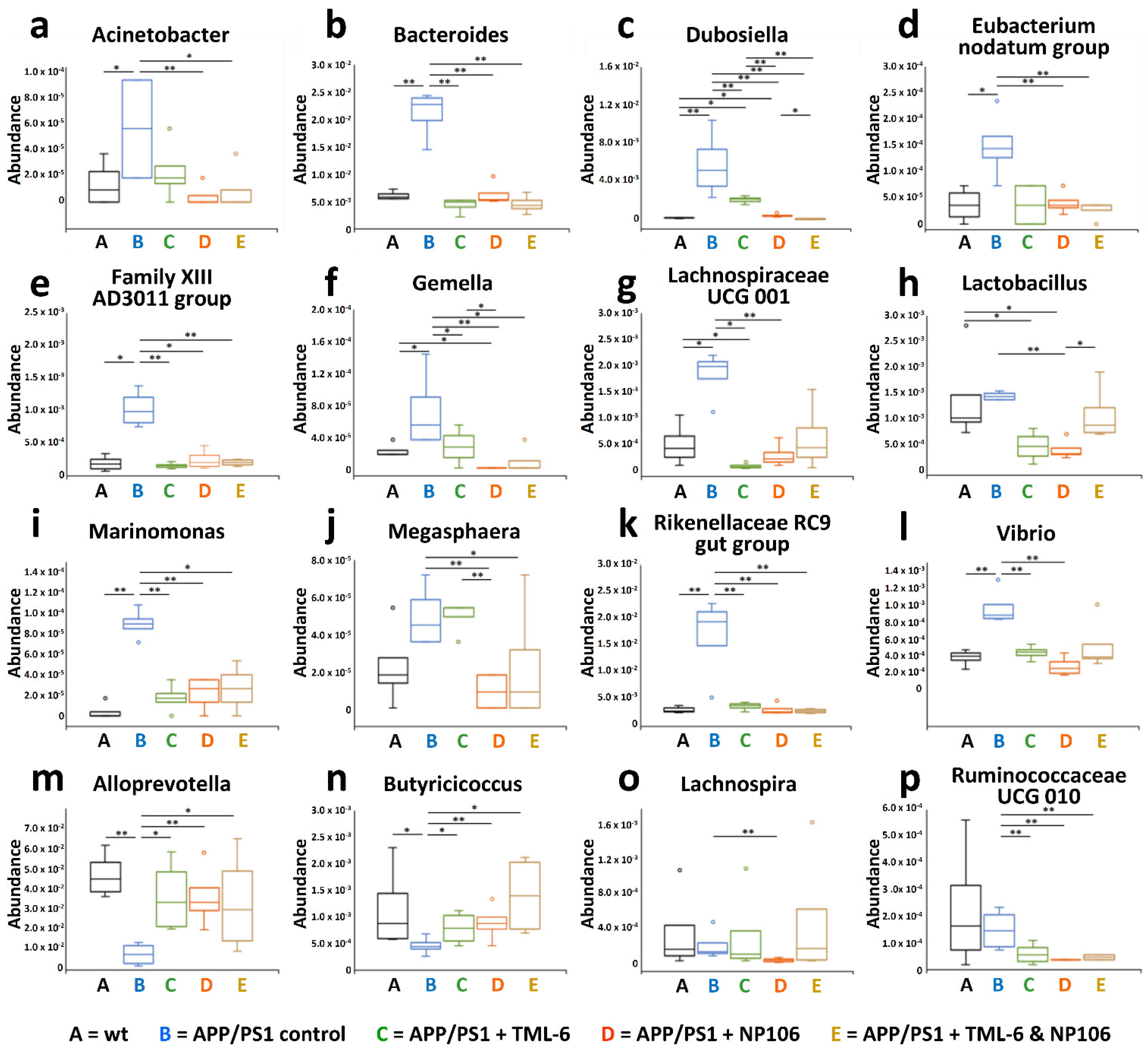
2.5. Combination Treatment Normalized Aberrant Bacterial Communities in APP/PS1 Mice to wt Levels
2.6. Associations of Abundances of Bacterial Genera with the Severity of Cerebral Aβ Pathology and Nesting Behavioral Abnormality
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Preparation of TML-6 Chow and Estimate of the Daily Uptake of TML-6
4.3. Treatment Regimens
4.4. Pathological and Morphological Examination Using Confocal Microscopy
4.5. Measurement of Levels of Aβ and Proinflammatory Cytokines Using ELISA
4.6. Nesting Behavioral Test
4.7. Gut Microbiota Analysis Using 16S rDNA Sequencing
4.8. Statistical Analyses
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Battaglia, S.; Garofalo, S.; di Pellegrino, G. Context-dependent extinction of threat memories: Influences of healthy aging. Sci. Rep. 2018, 8, 12592. [Google Scholar] [CrossRef]
- Serý, O.; Povová, J.; Míšek, I.; Pešák, L.; Janout, V. Molecular mechanisms of neuropathological changes in Alzheimer’s disease: A review. Folia Neuropathol. 2013, 51, 1–9. [Google Scholar] [CrossRef]
- Yan, R.; Vassar, R. Targeting the β secretase BACE1 for AD therapy. Lancet Neurol. 2014, 13, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Gregori, M.; Masserini, M.; Mancini, S. Nanomedicine for the treatment of AD. Nanomedicine 2015, 10, 1203–1218. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.; Marshall, G.A. Primary and secondary prevention trials in Alzheimer Disease: Looking back, moving forward. Curr. Alzheimer Res. 2017, 14, 426–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, V.H.; Nicoll, J.A.R.; Holmes, C. Microglia in neurodegenerative disease. Nat. Rev. Neurol. 2010, 6, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Michaud, J.P.; Rivest, S. Anti-inflammatory signaling in microglia exacerbates Alzheimer’s disease-related pathology. Neuron 2015, 85, 450–452. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Hong, J.S. Role of microglia in inflammation-mediated neurodegenerative diseases: Mechanisms and strategies for therapeutic intervention. J. Pharmacol. Exp. Ther. 2003, 304, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Dheen, S.T.; Kaur, C.; Ling, E.A. Microglial activation and its implications in the brain diseases. Curr. Med. Chem. 2007, 14, 1189–1197. [Google Scholar] [CrossRef]
- Rawji, K.S.; Mishra, M.K.; Michaels, N.J.; Rivest, S.; Stys, P.K.; Yong, V.W. Immunosenescence of microglia and macrophages: Impact on the ageing central nervous system. Brain 2016, 139, 653–661. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.T. Microglial aging in the healthy CNS: Phenotypes, drivers, and rejuvenation. Front. Cell Neurosci. 2013, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [PubMed]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Erny, D.; de Angelis, A.L.H.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Matcovitch-Natan, O.; Winter, D.R.; Giladi, A.; Aguilar, S.V.; Spinrad, A.; Sarrazin, S.; Ben-Yehuda, H.; David, E.; González, F.Z.; Perrin, P.; et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 2016, 353, aad8670. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Ling, Z.; Zhu, M.; Yan, X.; Cheng, Y.; Shao, L.; Liu, X.; Jiang, R.; Wu, S. Structural and functional dysbiosis of fecal microbiota in Chinese patients with Alzheimer’s disease. Front. Cell Dev. Biol. 2021, 8, 634069. [Google Scholar] [CrossRef]
- Sun, J.; Xu, J.; Yang, B.; Chen, K.; Kong, Y.; Fang, N.; Gong, T.; Wang, F.; Ling, Z.; Liu, J. Effect of Clostridium butyricum against microglia-mediated neuroinflammation in Alzheimer’s disease via regulating gut microbiota and metabolites butyrate. Mol. Nutr. Food Res. 2020, 64, e1900636. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Lee, G.; Ritter, A.; Zhong, K. Alzheimer’s disease drug development pipeline: 2018. Alzheimer’s Dement. 2018, 4, 195–214. [Google Scholar] [CrossRef]
- Geerts, H.; Spiros, A. Learning from amyloid trials in Alzheimer’s disease. A virtual patient analysis using a quantitative systems pharmacology approach. Alzheimer’s Dement. 2020, 16, 862–872. [Google Scholar] [CrossRef]
- VandeVrede, L.; Gibbs, D.M.; Koestler, M.; La Joie, R.; Ljubenkov, P.A.; Provost, K.; Soleimani-Meigooni, D.; Strom, A.; Tsoy, E.; Rabinovici, G.D.; et al. Symptomatic amyloid-related imaging abnormalities in an APOE ε4/ε4 patient treated with aducanumab. Alzheimers Dement. 2020, 12, e12101. [Google Scholar] [CrossRef] [PubMed]
- Fillit, H.; Green, A. Aducanumab and the FDA—where are we now? Nat. Rev. Neurol. 2021, 17, 129–130. [Google Scholar] [CrossRef] [PubMed]
- Knopman, D.S.; Jones, D.T.; Greicius, M.D. Failure to demonstrate efficacy of aducanumab: An analysis of the EMERGE and ENGAGE trials as reported by Biogen. Alzheimers Dement. 2021, 17, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef]
- Battaglia, S.; Serio, G.; Scarpazza, C.; D’Ausilio, A.; Borgomaneri, S. Frozen in (e)motion: How reactive motor inhibition is influenced by the emotional content of stimuli in healthy and psychiatric populations. Behav. Res. Ther. 2021, 146, 103963. [Google Scholar] [CrossRef]
- Ibrahim, M.M.; Gabr, M.T. Multitarget therapeutic strategies for Alzheimer’s disease. Neural Regen. Res. 2019, 14, 437–440. [Google Scholar]
- Sarlus, H.; Heneka, M.T. Microglia in Alzheimer’s disease. J. Clin. Investig. 2017, 127, 3240–3249. [Google Scholar] [CrossRef]
- Hsu, J.T.A.; Tien, C.F.; Yu, G.Y.; Shen, S.; Lee, Y.H.; Hsu, P.C.; Wang, Y.; Chao, P.K.; Tsay, H.J.; Shie, F.S. The effects of Aβ1–42 binding to the SARS-CoV-2 spike protein S1 subunit and angiotensin-converting enzyme 2. Int. J. Mol. Sci. 2021, 22, 8226. [Google Scholar] [CrossRef] [PubMed]
- Su, I.J.; Chang, H.Y.; Wang, H.C.; Tsai, K.J. A curcumin analog exhibits multiple biologic effects on the pathogenesis of AD and improves behavior, inflammation, and Aβ accumulation in a mouse model. Int. J. Mol. Sci. 2020, 21, 5459. [Google Scholar] [CrossRef]
- Deacon, R.M.J. Assessing nest building in mice. Nat. Protoc. 2006, 1, 1117–1119. [Google Scholar] [CrossRef]
- Filali, M.; Lalonde, R. Age-related cognitive decline and nesting behavior in an APPswe/PS1 bigenic model of Alzheimer’s disease. Brain Res. 2009, 1292, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Jing, D.; Liu, Z.; Chen, Y.; Huang, F.; Behnisch, T. Enhanced expression of secreted α-klotho in the hippocampus alters nesting behavior and memory formation in mice. Front. Cell Neurosci. 2019, 13, 133. [Google Scholar] [CrossRef]
- Etherton, M.R.; Blaiss, C.A.; Powell, C.M.; Südhof, T.C. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl. Acad. Sci. USA 2009, 106, 17998–18003. [Google Scholar] [CrossRef] [Green Version]
- Kondratiuk, I.; Devijver, H.; Lechat, B.; Van Leuven, F.; Kaczmarek, L.; Filipkowski, R.K. Glycogen synthase kinase-3beta affects size of dentate gyrus and species-typical behavioral tasks in transgenic and knockout mice. Behav. Brain Res. 2013, 248, 46–50. [Google Scholar] [CrossRef]
- Jedynak, P.; Jaholkowski, P.; Wozniak, G.; Sandi, C.; Kaczmarek, L.; Filipkowski, R.K. Lack of cyclin D2 impairing adult brain neurogenesis alters hippocampal-dependent behavioral tasks without reducing learning ability. Behav. Brain Res. 2012, 227, 159–166. [Google Scholar] [CrossRef]
- Saji, N.; Murotani, K.; Hisada, T.; Tsuduki, T.; Sugimoto, T.; Kimura, A.; Niida, S.; Toba, K.; Sakurai, T. The relationship between the gut microbiome and mild cognitive impairment in patients without dementia: A cross-sectional study conducted in Japan. Sci. Rep. 2019, 9, 19227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Peng, J.; Huang, X.; Xiao, L.; Huang, F.; Zuo, Z. Gut microbiome features of Chinese patients newly diagnosed with Alzheimer’s disease or mild cognitive impairment. J. Alzheimers Dis. 2021, 80, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, Y.; Ma, J.; Huang, P.; Du, J.; Cao, L.; Wang, Y.; Xiao, Q.; Tang, H.; Chen, S. Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement. 2019, 15, 1357–1366. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Machiels, K.; Perrier, C.; Romero, C.; Maes, S.; Flahou, B.; Steppe, M.; Haesebrouck, F.; Sas, B.; Ducatelle, R.; et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 2013, 62, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Borgomaneri, S.; Battaglia, S.; Garofalo, S.; Tortora, F.; Avenanti, A.; di Pellegrino, G. State-Dependent TMS over Prefrontal Cortex Disrupts Fear-Memory Reconsolidation and Prevents the Return of Fear. Curr. Biol. 2020, 30, 3672–3679. [Google Scholar] [CrossRef]
- Borgomaneri, S.; Battaglia, S.; Sciamanna, G.; Tortora, F.; Laricchiuta, D. Memories are not written in stone: Re-writing fear memories by means of non-invasive brain stimulation and optogenetic manipulations. Neurosci. Biobehav. Rev. 2021, 127, 334–352. [Google Scholar] [CrossRef]
- Borgomaneri, S.; Battaglia, S.; Avenanti, A.; di Pellegrino, G. Corrigendum to Don’t Hurt Me No More: State-dependent Transcranial Magnetic Stimulation for the treatment of specific phobia. J. Affect. Disord. 2021, 286, 78–79. [Google Scholar] [CrossRef]
- Lynch, C.J.; Breeden, A.L.; Gordon, E.M.; Cherry, J.B.C.; Turkeltaub, P.E.; Vaidya, C.J. Precision Inhibitory Stimulation of Individual-Specific Cortical Hubs Disrupts Information Processing in Humans. Cereb. Cortex. 2019, 29, 3912–3921. [Google Scholar] [CrossRef]
- Roshchupkina, L.; Stee, W.; Peigneux, P. Beta-tACS does not impact the dynamics of motor memory consolidation. Brain Stimul. 2020, 13, 1489–1490. [Google Scholar] [CrossRef]
- Vosskuhl, J.; Strüber, D.; Herrmann, C.S. Non-invasive Brain Stimulation: A Paradigm Shift in Understanding Brain Oscillations. Front. Hum. Neurosci. 2018, 12, 211. [Google Scholar] [CrossRef] [Green Version]
- Ferreri, F.; Guerra, A.; Vollero, L.; Ponzo, D.; Määtta, S.; Könönen, M.; Vecchio, F.; Pasqualetti, P.; Miraglia, F.; Simonelli, I.; et al. TMS-EEG Biomarkers of Amnestic Mild Cognitive Impairment Due to Alzheimer’s Disease: A Proof-of-Concept Six Years Prospective Study. Front. Aging Neurosci. 2021, 13, 737281. [Google Scholar] [CrossRef] [PubMed]
- Sprugnoli, G.; Munsch, F.; Cappon, D.; Paciorek, R.; Macone, J.; Connor, A.; El Fakhri, G.; Salvador, R.; Ruffini, G.; Donohoe, K.; et al. Impact of multisession 40 Hz tACS on hippocampal perfusion in patients with Alzheimer’s disease. Alzheimers Res. Ther. 2021, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.W.; Yeh, S.H.; Shie, F.S.; Lai, W.S.; Liu, H.K.; Tzeng, T.T.; Tsay, H.J.; Shiao, Y.J. Impaired cognition and cerebral glucose regulation are associated with astrocyte activation in the parenchyma of metabolically stressed APPswe/PS1dE9 mice. Neurobiol. Aging 2015, 36, 2984–2994. [Google Scholar] [CrossRef] [PubMed]




| Anosim | MRPP | Adonis | ||||
|---|---|---|---|---|---|---|
| Group | R | p Value | Expected δ | p Value | R2 | p Value |
| A vs. B | 1 | 0.028 * | 0.42 | 0.041 * | 0.53 | 0.026 * |
| A vs. C | 0.875 | 0.026 * | 0.34 | 0.024 * | 0.36 | 0.028 * |
| A vs. D | 0.490 | 0.049 * | 0.36 | 0.034 * | 0.27 | 0.057 |
| A vs. E | 0.281 | 0.124 | 0.35 | 0.103 | 0.22 | 0.140 |
| B vs. C | 1 | 0.029 * | 0.44 | 0.030 * | 0.62 | 0.028 * |
| B vs. D | 0.906 | 0.031 * | 0.43 | 0.035 * | 0.48 | 0.028 * |
| B vs. E | 0.938 | 0.031 * | 0.44 | 0.037 * | 0.49 | 0.028 * |
| C vs. D | 0.865 | 0.026 * | 0.39 | 0.022 * | 0.43 | 0.026 * |
| C vs. E | 0.438 | 0.024 * | 0.37 | 0.027 * | 0.36 | 0.030 * |
| D vs. E | 0.104 | 0.194 | 0.36 | 0.224 | 0.18 | 0.223 |
| Nesting | Area of Aβ Plaques | Number of Aβ Plaques | Aβ Plaques >500 μm2 | Aβ Plaques <500 μm2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | p Value | R | p Value | R | p Value | R | p Value | R | p Value | |
| Genera Abundance Increased in APP/PS1 Mice | ||||||||||
| Acinetobacter | −0.493 | 0.027 * | 0.363 | 0.167 | 0.415 | 0.110 | 0.283 | 0.288 | 0.419 | 0.106 |
| Bacteroides | −0.503 | 0.024 * | 0.607 | 0.013 * | 0.586 | 0.017 * | 0.583 | 0.018 * | 0.550 | 0.027 * |
| Dubosiella | −0.756 | < 0.001 *** | 0.543 | 0.030 * | 0.649 | 0.007 ** | 0.331 | 0.210 | 0.680 | 0.004 ** |
| Eubacterium nodatum group | −0.761 | < 0.001 *** | 0.505 | 0.046 * | 0.500 | 0.049 * | 0.411 | 0.114 | 0.488 | 0.055 |
| Family XIII AD3011 group | −0.526 | 0.017 * | 0.590 | 0.016 * | 0.592 | 0.016 * | 0.570 | 0.021 * | 0.560 | 0.024 * |
| Gemella | −0.412 | 0.071 | 0.585 | 0.017 * | 0.541 | 0.030 * | 0.518 | 0.040 * | 0.512 | 0.042 * |
| Lachnospiraceae UCG 001 | −0.164 | 0.491 | 0.477 | 0.062 | 0.416 | 0.109 | 0.471 | 0.065 | 0.377 | 0.149 |
| Lactobacillus | −0.064 | 0.789 | 0.278 | 0.316 | 0.313 | 0.238 | 0.181 | 0.502 | 0.323 | 0.221 |
| Marinomonas | −0.626 | 0.003 ** | 0.598 | 0.014 * | 0.647 | 0.007 ** | 0.463 | 0.071 | 0.649 | 0.007 ** |
| Megasphaera | −0.425 | 0.062 | 0.339 | 0.199 | 0.415 | 0.110 | 0.263 | 0.325 | 0.424 | 0.102 |
| Rikenellaceae RC9 gut group | −0.691 | < 0.001 *** | 0.592 | 0.030 * | 0.502 | 0.048 * | 0.536 | 0.032 * | 0.462 | 0.072 |
| Vibrio | −0.363 | 0.116 | 0.391 | 0.134 | 0.472 | 0.065 | 0.264 | 0.324 | 0.490 | 0.054 |
| Genera abundance decreased in APP/PS1 mice | ||||||||||
| Alloprevotella | 0.320 | 0.169 | −0.080 | 0.769 | −0.154 | 0.569 | −0.060 | 0.825 | −0.168 | 0.535 |
| Butyricicoccus | 0.622 | 0.003 ** | −0.509 | 0.044 * | −0.546 | 0.029 * | −0.433 | 0.094 | −0.536 | 0.032 * |
| Lachnospira | 0.150 | 0.528 | −0.241 | 0.369 | −0.121 | 0.656 | −0.327 | 0.217 | −0.066 | 0.808 |
| Ruminococcaceae UCG 010 | −0.163 | 0.493 | 0.381 | 0.145 | 0.398 | 0.127 | 0.343 | 0.193 | 0.383 | 0.142 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, I.-J.; Hsu, C.-Y.; Shen, S.; Chao, P.-K.; Hsu, J.T.-A.; Hsueh, J.-T.; Liang, J.-J.; Hsu, Y.-T.; Shie, F.-S. The Beneficial Effects of Combining Anti-Aβ Antibody NP106 and Curcumin Analog TML-6 on the Treatment of Alzheimer’s Disease in APP/PS1 Mice. Int. J. Mol. Sci. 2022, 23, 556. https://doi.org/10.3390/ijms23010556
Su I-J, Hsu C-Y, Shen S, Chao P-K, Hsu JT-A, Hsueh J-T, Liang J-J, Hsu Y-T, Shie F-S. The Beneficial Effects of Combining Anti-Aβ Antibody NP106 and Curcumin Analog TML-6 on the Treatment of Alzheimer’s Disease in APP/PS1 Mice. International Journal of Molecular Sciences. 2022; 23(1):556. https://doi.org/10.3390/ijms23010556
Chicago/Turabian StyleSu, Ih-Jen, Chia-Yu Hsu, Santai Shen, Po-Kuan Chao, John Tsu-An Hsu, Jung-Tsung Hsueh, Jia-Jun Liang, Ying-Ting Hsu, and Feng-Shiun Shie. 2022. "The Beneficial Effects of Combining Anti-Aβ Antibody NP106 and Curcumin Analog TML-6 on the Treatment of Alzheimer’s Disease in APP/PS1 Mice" International Journal of Molecular Sciences 23, no. 1: 556. https://doi.org/10.3390/ijms23010556






